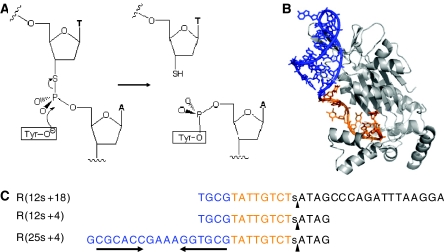
Figure 2.
Cleavage reaction of phosphorothiolate oligonucleotides. (A) Scheme of TrwC cleavage reaction. In the normal reaction, the hydroxyl group of the catalytic tyrosil residue is a nucleophile that attacks the phosphate group in the nic site. As a consequence, the phosphodiester bond is broken and a free 3′-OH group is generated. When the oxygen in the scissile P–O bond is replaced by sulfur, the resulting cleavage product ends in a 3′-SH group. The religation reaction that reverses the cleavage reaction when normal oligonucleotides are used is thus inhibited with phosphorothiolate oligonucleotides. As a consequence, the equilibrium is displaced towards oligonucleotide–protein covalent complex formation. (B) X-Ray structure of TrwCR in complex with DNA (pdb:1OMH). The color scheme of the DNA reflects the double- (blue) or single-stranded (ochre) DNA portions of the bound oligonucleotide, and parallels the colors of the sequences in (C). (C) Nucleotide sequences of oligonucleotides R(12s+18), R(12s+4) and R(25s+4), used in this study. The position of the phosphorothiolate modification is shown by a lower case s within the sequences. The cleavage sites are represented by black vertical arrowheads. The 25 nucleotide-long 5′-moiety of R(25s+4) contains an inverted repeat sequence (horizontal arrows) linked via a GAAA loop that results in the double stranded DNA colored blue in (B).