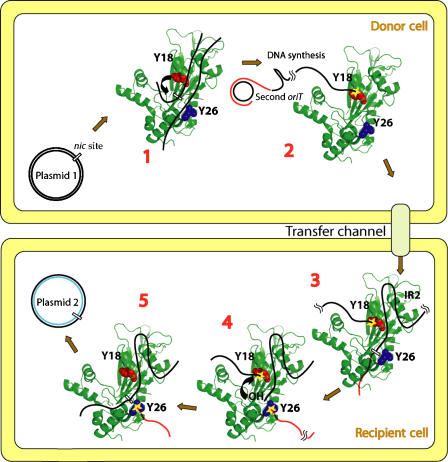
Figure 6.
Model for the mechanism of DNA processing during R388 plasmid conjugation. (1) First oriT binding: TrwC (the protein structure is represented as a cartoon, colored in green; Y18 and Y26 residues are shown as spacefill representations in red and blue, respectively) binds supercoiled R388 plasmid DNA (black lines) around the nic site in oriT. (2) Y18 cleavage: TrwC tyrosine Y18 cleaves R388 at nic (curved arrow in (1)). Upon cleavage, TrwC becomes bound to the 5′ end of nic (yellow star). RCR in the donor plasmid DNA (new DNA represented as a red line) displaces the T-strand. TrwC bound to the T-strand pilots the DNA to the recipient cell by the transfer channel. (3) Second oriT binding: TrwC recognizes IR2 when the trailing DNA arrives in the recipient cell and specifically binds nic of the second oriT. (4) Y26 cleavage: TrwC tyrosine Y26 cleaves the second oriT at nic, forming a second phosphotyrosine bond (yellow star bound to red line). A free 3′-OH is generated that attacks the Y18–DNA complex (curved arrow in (4)). (5) Plasmid release: DNA-strand ligation occurs by a second transesterification event, when the newly generated 3′-OH cleaves the phosphotyrosyl bond and the covalent intermediate is resolved. This last step allows recircularization and release of the transferred plasmid DNA.