A homo-tetrameric psychrophilic alcohol dehydrogenase from the Antarctic psychrophile Moraxella sp. TAE123 has been crystallized. Its molecular structure is under investigation.
Keywords: alcohol dehydrogenases, psychrophilic, NAD(H)-dependent, Moraxella sp. TAE123
Abstract
An NAD+-dependent psychrophilic alcohol dehydrogenase (ADH) from the Antarctic psychrophile Moraxella sp. TAE123 has been purified to homogeneity. The enzyme consists of four identical subunits, each containing two Zn ions. Protein crystals suitable for X-ray diffraction were obtained under optimized salting-out crystallization conditions using ammonium sulfate as a precipitating agent. The crystals are hexagonal bipyramids and belong to space group P3121 or P3221, with unit-cell parameters a = 136.4, c = 210.7 Å. They contain one protein homotetramer in the asymmetric unit. Diffraction data were collected to 2.2 Å under cryogenic conditions using synchrotron radiation.
1. Introduction
Most of the cold-adapted enzymes that have been characterized to date originate from organisms isolated from the Antarctic and the Antarctic seawater. In these low-temperature environments the rates of the chemical reactions responsible for maintaining life are substantially reduced. Despite this, organisms native to cold environments achieve metabolic rates that are sufficient for survival and growth and comparable with those exhibited by mesophilic organisms at moderate temperatures. These organisms synthesize enzymes characterized by a high catalytic activity at low temperatures associated, however, with low thermal stability (Feller, 2003 ▶).
Alcohol dehydrogenases (ADHs; EC 1.1.1.1) belong to the oxidoreductase family, a class of enzymes responsible for the catalysis of all biological oxidation–reduction reactions. Alcohol dehydrogenases catalyse the cofactor-dependent interconversion of alcohols to their corresponding aldehydes or ketones. The enzymes are widely distributed and are present throughout the three domains of life: archaea, bacteria and eukarya (Rella et al., 1987 ▶; Von Wartburg et al., 1964 ▶). The NAD(P)+-dependent ADHs may be separated into three distinct classes based on molecular size (Reid & Fewson, 1994 ▶). The type I or medium-chain ADHs form the largest and most extensively studied group. With approximately 370 residues per chain, this class includes the well characterized horse liver ADH (Eklund et al., 1976 ▶). Enzymes of this class may be dimeric or tetrameric and all contain bound zinc ions. In general, bacterial type I ADHs are tetramers, while those from higher eukaryotes are dimeric enzymes. Type II ADHs are classified as short chain, having approximately 250 residues per chain, and do not normally contain metals (Persson et al., 1991 ▶). This group includes the ADH found in Drosophila melanogaster (Thatcher, 1980 ▶). Type III or long-chain ADHs contain over 380 but often as many as 900 amino-acid residues. Enzymes of this class are frequently activated by iron. Represented by ADH II from Zymomonas mobilis (Scopes, 1983 ▶), these form the least characterized class and show little sequence similarity to type I or type II ADHs.
Crystal structures are available for a number of eukaryotic and prokaryotic type I ADHs. Horse liver ADH was the first to be studied by X-ray crystallography (Eklund et al., 1976 ▶); more recently, an atomic resolution structure has been obtained (Meijers et al., 2001 ▶). Three ADHs have been crystallized from thermophiles. The NADP(H)-dependent ADH from the thermophilic bacterium Thermoanaerobium brockii was crystallized in its holoenzyme form and the structure was solved to 2.5 Å resolution (Korkhin et al., 1998 ▶). The X-ray structure of a tetrameric NAD(H)-dependent ADH from the hyperthermophilic archaeon Sulfolobus solfataricus has been solved in its apo form (Esposito et al., 2002 ▶) and more recently as a ternary complex with NAD(H) and 2-ethoxyethanol (Esposito et al., 2003 ▶). Finally, a tetrameric type I ADH from the archaeon Aeropyrum pernix has been solved recently with NADH and octanoic acid present at the active site of the enzyme (Guy et al., 2003 ▶).
In the present work, we report the crystallization and preliminary X-ray data analysis of the first cold-adapted ADH from the psychrophile Moraxella sp. TAE123. This ADH is a type I ADH and exhibits a wide range of substrate specificity, oxidizing mainly primary and secondary aliphatic alcohols utilizing NAD+ as coenzyme. It shows highest reaction rates for ethanol as a substrate and gradually decreases its reaction rates as the length and branching of the carbon chain of the alcohol substrates increases. The enzyme imposes effective reduction of aldehydes and ketones. The present psychrophilic ADH shares 69 and 60% sequence identity with the known ADH structures from Pseudomonas aeruginosa (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1llu) and Bacillus stearothermophilus (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1rjw), respectively. It shows 25–35% sequence identity with ADHs from eukaryotes and mesophilic and thermophilic bacteria. It also exhibits significant sequence identity (74–62%), in decreasing order, with ADHs from the bacteria Brucella suis, B. melitensis, Acinetobacter sp., Ralstonia solanacearum, Burkholderia mallei, Cytophaga sp., Agrobacterium tumefaciens and Rhizobium meliloti. The active psychrophilic enzyme is homo-tetrameric and consists of four subunits, each containing 346 amino-acid residues (Tsigos et al., manuscript in preparation). It has a molecular weight of 146 932 Da as deduced from the amino-acid sequence.
2. Materials and methods
2.1. Expression and purification
The ADH gene was isolated from a genomic expression library of Moraxella sp. TAE123. The gene was subcloned in the pET-26b(+) (Novagen) expression vector. The resulting pET-26b(+)-ADH plasmid was transformed into Escherichia coli BL21(DE3) for expression of the ADH C-terminal 6×His-tagged fusion protein. The transformed cells were grown at 297 K in 2 l LB growth medium supplemented with 0.25 mM ZnSO4 and 30 µg ml−1 kanamycin. The culture was induced for expression of ADH by the addition of 0.3 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at an absorbance of OD600 = 1. The culture was centrifuged after 12 h of induction at 297 K. The cell paste obtained was suspended in 20 ml lysis buffer A (50 mM Tris pH 8.0, 200 mM NaCl) in the presence of 1 mM phenylmethylsulfonyl fluoride (PMSF) and lysed using a French press. The lysate was centrifuged for 60 min at 15 000g and 279 K and the supernatant was mixed with 5 ml Ni–NTA (Qiagen) beads. The suspension was shaken slowly overnight at 277 K. All subsequent steps were performed at 293 K. The Ni–NTA beads were thoroughly washed four times with 50 ml buffer A. The washed beads were packed into a chromatographic column and washed with ten bed volumes of buffer B (50 mM Tris pH 8.0, 5 mM imidazole) followed by ten bed volumes of buffer B containing 1 M NaCl and once again with ten bed volumes of buffer B containing 200 mM NaCl. The protein was eluted from the chelating affinity resin by step elution with 200 mM imidazole and was concentrated to 2 ml. The concentrated protein was loaded onto a 220 ml Sephacryl S-300 High Resolution (Pharmacia) gel-filtration column, which was equilibrated with 20 mM Tris pH 7.0, 200 mM NaCl. ADH elutes mainly as a tetrameric protein (4 × 36 733 Da). A smaller amount of the enzyme elutes as a monomeric inactive protein. The purity of the sample was assessed by denaturing polyacrylamide gel electrophoresis (Laemmli, 1970 ▶). Finally, the protein solution was extensively dialysed against storage buffer (5 mM Tris pH 7.0 and 100 mM NaCl) and concentrated to 20 mg ml−1 with an Ultrafree-15 centrifugal filter device (Millipore). The protein concentration was determined photometrically as described by Bradford (1976 ▶). The concentrated protein solution was divided into 250 µl aliquots and frozen at 253 K.
2.2. Crystallization
In the initial crystallization trials, the hanging-drop vapour-diffusion method was used with droplets consisting of 1 µl protein solution and 1 µl reservoir solution. Several salting-out crystallization conditions were identified using ammonium sulfate as the precipitating agent. The best quality crystals were obtained at 291 K using a hanging droplet that contained 6 µl protein aliquot and 2 µl reservoir solution (100 mM Bis-Tris pH 7.0, 2 M ammonium sulfate, 100 mM NaCl, 100 mM imidazole). The reservoir solution was placed on top of the protein solution without mixing. Imidazole greatly improves the crystal quality and size. Crystals grew to their final size in a period of one week.
2.3. Crystallographic data collection
Initially, data were collected in-house at both 290 and 100 K using a MAR300 (MAR Research, Hamburg) image-plate detector and Cu Kα radiation from a Rigaku RU-3HR rotating-anode X-ray generator. High-resolution data were collected from a single crystal at the European Synchrotron Radiation Facility (ESRF) beamline ID14-1 using a Quantum 4R (ADSC) detector and radiation of wavelength 0.934 Å at 100 K. The cryoprotection solution contained 100 mM Bis-Tris pH 7.0, 2 M ammonium sulfate, 100 mM NaCl and 20%(v/v) glycerol. The crystals were very sensitive to changes in the mother liquor, exhibiting loss of diffracting power. In order to avoid this behaviour, the crystal to be measured was washed for a few seconds in the cryoprotection solution and immediately plunged into liquid nitrogen before mounting on the goniometer head.
3. Results and discussion
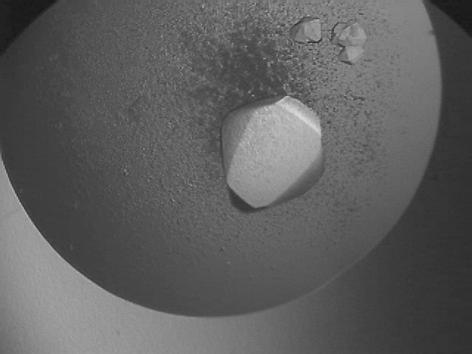
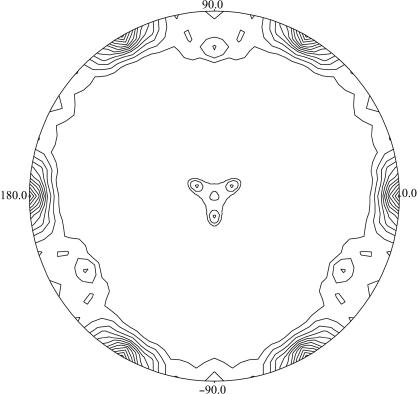
Single hexagonal bipyramidal crystals were obtained in approximately one week with maximum dimensions of about 0.4 × 0.4 × 1 mm (Fig. 1 ▶). The crystals diffracted to better than 2.5 Å using synchrotron radiation and belonged to the trigonal space group P3121 or its enantiomorph P3221, with unit-cell parameters a = b = 136.4, c = 210.7 Å. A single set of diffraction data to 2.2 Å resolution was collected at a high-energy remote wavelength for Zn (0.934 Å) from one very large crystal (1.5 mm long). The data were processed using the programs MOSFLM (Leslie, 1992 ▶) and SCALA (Collaborative Computational Project, Number 4, 1994 ▶). A summary of the X-ray diffraction data statistics is shown in Table 1 ▶. The solvent content of the crystals has been estimated at 68%(v/v) and the V M ratio as 3.85 Å3 Da−1 (Matthews, 1968 ▶). The latter value is higher than the previously reported V M values (2.5–3.1 Å3 Da−1) for crystals of type I ADHs. This fact could be assigned to the different crystal packing. The asymmetric unit contains an enzyme homotetramer (4 × 346 amino-acid residues). A self-rotation function (Fig. 2 ▶) suggests that the ADH tetramer exhibits 222 point-group symmetry. Two of the molecular dyads are local symmetry axes and one is parallel to the crystallographic rotational twofold axis. The diffraction data have been phased successfully using the relatively small anomalous signal from the Zn2+ ions (2.8% average intensity difference within the Bijvoet pairs of reflections). The programs SHELXD (Schneider & Sheldrick, 2002 ▶) and SHARP (de La Fortelle & Bricogne, 1997 ▶) were used to locate the anomalous scatterers and to refine them, respectively. The value of the initial overall figure of merit was 0.265 for the entire range of data. The molecular arrangement based on the self-rotation function has been confirmed from the derived model from the initial set of phases. The refinement of the enzyme structure is in progress.
Figure 1.
Truncated hexagonal bipyramidal crystals of Moraxella sp. TAE123 psychrophilic ADH. The longest dimension of the large crystal is approximately 1 mm.
Table 1. Crystal parameters and processing statistics for the synchrotron data set.
Values in parentheses are for the highest resolution shell.
| Space group | P3121 or P3221 |
| Unit-cell parameters (Å) | a = b = 136.4, c = 210.7 |
| Wavelength (Å) | 0.934 |
| Resolution range (Å) | 25–2.2 (2.32–2.2) |
| No. of measured reflections | 1373149 (139948) |
| No. of unique reflections† | 115606 (16711) |
| Completeness (%) | 99.9 (100.0) |
| Multiplicity | 11.9 (8.4) |
| Anomalous completeness (%) | 99.9 (99.9) |
| Anomalous multiplicity | 6.0 (4.2) |
| 〈I〉/σ〈I〉 | 18.9 (4.3) |
| Rsym‡ | 0.078 (0.438) |
I + and I − were scaled separately.
R
sym = 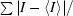
 , where I is the intensity of reflection.
, where I is the intensity of reflection.
Figure 2.
Self-rotation function of the Moraxella sp. TAE123 psychrophilic ADH, κ = 180° section. Radial coordinate, ω; angular coordinate, ϕ. The function was calculated with reflections in the resolution range 14–3 Å and an integration radius of 18 Å using the program POLARRFN from the CCP4 program package (Collaborative Computational Project, Number 4, 1994 ▶). Contouring of the map starts at the 2σ level with steps of 0.5σ. Two molecular dyads of the ADH tetramer are visible at polar angles (ω, ϕ) of (77, 90°) and (13, 270°), while the third molecular dyad is parallel to the crystallographic x axis.
Acknowledgments
We would like to thank the EMBL Grenoble Outstation for providing support for measurements at the ESRF under the European Community Research Infrastructure Action FP6 program.
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Eklund, H., Nordstrom, B., Zeppezauer, E., Soderlund, G., Ohlsson, I., Boiwe, T., Soderberg, B., Tapia, O. & Branden, C. I. (1976). J. Mol. Biol.102, 27–59. [DOI] [PubMed] [Google Scholar]
- Esposito, L., Bruno, I., Sica, F., Raia, C. A., Giordano, A., Rossi, M., Mazzarella, L. & Zagari, A. (2003). Biochemistry, 42, 14397–14407. [DOI] [PubMed] [Google Scholar]
- Esposito, L., Sica, F., Raia, C. A., Giordano, A., Rossi, M., Mazzarella, L. & Zagari, A. (2002). J. Mol. Biol.318, 463–477. [DOI] [PubMed] [Google Scholar]
- Feller, G. (2003). Cell. Mol. Life Sci.60, 648–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy, J. E., Isupov, M. N. & Littlechild, J. A. (2003). J. Mol. Biol.331, 1041–1051. [DOI] [PubMed] [Google Scholar]
- Korkhin, A., Kalb (Gilboa), J., Peretz, M., Bogin, O., Burstein, Y. & Frolow, F. (1998). J. Mol. Biol.278, 967–981. [DOI] [PubMed] [Google Scholar]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed] [Google Scholar]
- La Fortelle, E. de & Bricogne, G. (1997). Methods Enzymol.276, 472–494. [DOI] [PubMed]
- Leslie, A. G. W. (1992). Jnt CCP4/ESF–EACBM Newsl. Protein Crystallogr.26
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Meijers, R., Morris, R. J., Adolph, H. W., Merli, A., Lamzin, V. S. & Cedergren-Zeppezauer, E. S. (2001). J. Biol. Chem.276, 9316–9321. [DOI] [PubMed] [Google Scholar]
- Persson, B., Krook, M. & Jornvall, H. (1991). Eur. J. Biochem.200, 537–543. [DOI] [PubMed] [Google Scholar]
- Reid, M. F. & Fewson, C. A. (1994). Crit. Rev. Microbiol.20, 13–56. [DOI] [PubMed] [Google Scholar]
- Rella, R., Raia, C. A., Pensa, M., Pisani, F. M., Gambacorta, A., De Rosa, M. & Rossi, M. (1987). Eur. J. Biochem.167, 475–479. [DOI] [PubMed] [Google Scholar]
- Schneider, T. R. & Sheldrick, G. M. (2002). Acta Cryst. D58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- Scopes, R. K. (1983). FEBS Lett.156, 303–306. [DOI] [PubMed] [Google Scholar]
- Thatcher, D. R. (1980). Biochem. J.187, 875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Wartburg, J. P., Bethune, J. L. & Vallee, B. L. (1964). Biochemistry, 3, 1775–1782. [DOI] [PubMed]