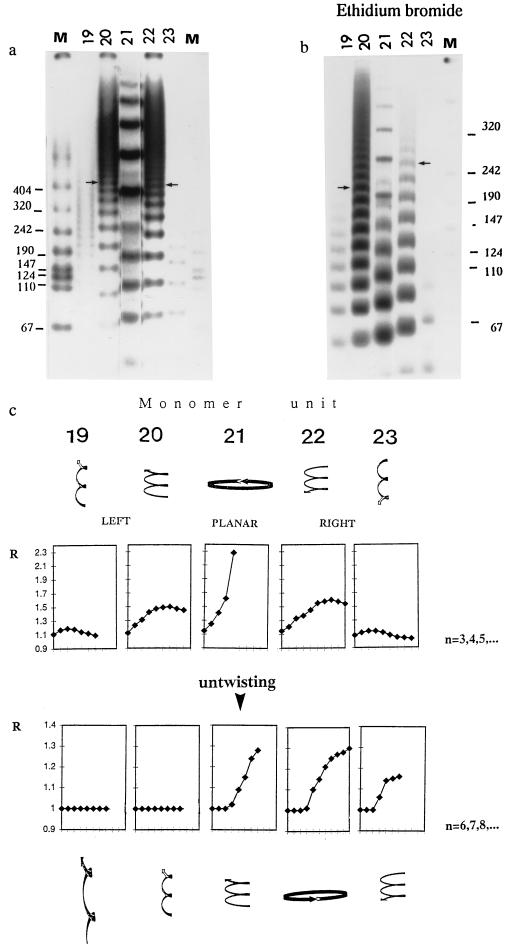
Figure 1.
Influence of untwisting on the gel mobility anomaly in curved DNA molecules (19- to 23-mers) with well-defined 3D shape: (a) Gel mobility under stress-free conditions (no EB). Monomer units used in ligations (19- to 23-mers) are designated at the top of each gel lane. Lane M is the molecular weight marker VII (Boehringer Mannheim). Arrows denote the similar mobilities of 220-bp left-handed and right-handed curved DNA fragments. (b) Gel mobility under untwisting conditions. Electrophoresis was done in the presence of 0.005 mg/ml of EB in the gel and gel-running buffer. Arrows denote the now quite different mobilities of 220-bp fragments of left-handed and right-handed curved DNA segments. (c) Gel mobility anomaly R versus n for the different monomer units (19- to 23-mers). R is the coefficient of retardation (13). The experimental error in determination of R is estimated as ±0.03. The number of ligated monomer units is denoted by “n.” Note that n starts at 3 (Upper) without untwisting and at 6 (Lower) with untwisting. Note also that the vertical scales of R are different for stress-free versus untwisting conditions. Highly schematized 3D shapes of curved DNA before and after untwisting are depicted at the top and bottom, respectively. Upon untwisting, the right-handed superhelixes (Right) increase their radii but decrease their overall twisting and become more planar. Initially planar curved DNA molecules (Center) are converted to left-handed superhelixes, while left-handed superhelixes (Left) become more twisted, with smaller radii.