The water-forming flavoenzyme NADH oxidase was crystallized successfully for the first time. The crystals diffract X-rays to at least 4.0 Å resolution.
Keywords: NADH oxidase, flavoenyzmes
Abstract
NADH oxidase (NOX) from Lactobacillus brevis is a homotetrameric flavoenzyme composed of 450 amino acids per subunit. The molecular weight of each monomer is 48.8 kDa. The enzyme catalyzes the oxidation of two equivalents of NADH and reduces one equivalent of oxygen to yield two equivalents of water, without releasing hydrogen peroxide after the reduction of the first equivalent of NADH. Crystals of this protein were grown in the presence of 34% polyethylene glycol monomethyl ether 2000, 0.1 M sodium acetate and 0.2 M ammonium sulfate at pH 5.4. They belong to the tetragonal space group P43212, with unit-cell parameters a = 74.8, b = 95.7, c = 116.9 Å, α = γ = 90, β = 103.8°. The current diffraction limit is 4.0 Å. The self-rotation function of the native data set is consistent with a NOX tetramer in the asymmetric unit.
1. Introduction
NADH oxidase (NOX) from Lactobacillus brevis is a homotetrameric flavoenzyme with a molecular weight of 48.8 kDa and one non-covalently bound FAD molecule per subunit. It catalyzes the oxidation of NADH by molecular oxygen to yield NAD+ and water without releasing hydrogen peroxide,
Quite recently, we overexpressed NOX from L. brevis and demonstrated its applicability for the regeneration of NAD+ (Hummel & Riebel, 2003 ▶, 2005 ▶). This type of NAD+ regeneration is useful for the application of NAD+-dependent dehydrogenases in oxidative processes, which resolve racemic mixtures of hydroxy acids, amino acids or alcohols, yielding enantiomerically pure compounds. The complete removal of one isomer is necessary to achieve a high enantiomeric excess of the product, which is strongly supported by the irreversibility of the regeneration reaction and the low K m value (18 µM) of the NADH oxidase from L. brevis.
This flavoprotein can be classified as an H2O-producing NADH oxidase. In addition to this NOX type of NADH oxidase, there are also H2O2-producing NADH oxidases such as the NADH oxidases from Thermus thermophilus, Sporolactobacillus inulinus (Park et al., 1992 ▶; Sakamoto & Komagata, 1996 ▶) and NADH peroxidases from lactic acid bacteria that reduce H2O2 to H2O (Parsonage & Claiborne, 1995 ▶; Sakamoto et al., 1996 ▶).
All these flavoenzymes may facilitate the regeneration of oxidized pyridine nucleotides for glycolysis and help to protect the organism against oxidative stress. For example, the NADH oxidases from Pyrococcus furiosus and L. delbrueckii have been shown to contribute significantly to aerobic metabolism under conditions of high O2 stress (Ward et al., 2001 ▶; Marty-Teysset et al., 2000 ▶). Higuchi and coworkers studied NADH oxidases from Streptococcus mutans, which are involved in energy metabolism and protection of the organism against oxidative stress (Higuchi et al., 1999 ▶).
Although several water-forming NADH oxidases have been characterized and described in the literature (Schmidt et al., 1986 ▶; Peterson et al., 1993 ▶; Kawasaki et al., 2004 ▶), no crystal structure of an NOX has been reported so far. We therefore present here the crystallization of such an NADH oxidase as the first step of its structure determination.
2. Materials and methods
2.1. Protein preparation
The nox gene of L. brevis was cloned and expressed in Escherichia coli by Hummel et al. (2003 ▶). E. coli Tuner(DE3) cells containing pET21a (Novagen) with nox were resuspended in 50 mM potassium phosphate buffer pH 6.5 (buffer A) at a ratio of 1 g cells (wet weight) to 3 ml buffer. Cells were disrupted by ultrasonification, cell debris was removed by centrifugation and the clear supernatant was loaded onto a column packed with Q-Sepharose FF (height 7.5 cm, diameter 1.6 cm; Amersham). This column was equilibrated with buffer A. After application of the enzyme sample, the column was washed with buffer A at a flow rate of 1.5 ml min−1 and NOX was eluted by application of a linear gradient of 0–1 M NaCl in buffer A. The active fractions were pooled, ammonium sulfate was added to a final concentration of 1.2 M and the pool was applied to a phenyl Sepharose FF column (height 7.5 cm, diameter 2.6 cm; Amersham). This second column was equilibrated with buffer A containing 1.2 M ammonium sulfate at a flow rate of 1.5 ml min−1. After washing, step elution was performed by decreasing the ammonium sulfate concentration using a linear gradient. The purity of protein samples and expression levels of NOX were analyzed using the NuPAGE system with 4–12% bis-Tris gels (Invitrogen).
Samples gained from this purification procedure and used for crystallization studies had a purity of >95% as judged by SDS–PAGE. MALDI mass spectrometry gave a molecular weight of 48.8 kDa for one subunit. This is in good correspondence with the theoretical molecular weight of 48 399 Da. Gel-filtration analysis showed an apparent molecular weight of 196 kDa, which corresponds to the weight of a tetramer. Prior to crystallization, the protein was concentrated and dialyzed against 10 mM MES pH 6.5 by ultrafiltration (Amicon). The final concentration of the protein was determined by the method of Bradford (1976 ▶) using bovine serum albumin as standard. The specific activity of L. brevis NOX was typically 350 U mg−1.
2.2. Crystallization
Initial crystallization experiments were based on the sparse-matrix sampling method (Jancarik & Kim, 1991 ▶). Crystals of NOX could be obtained at pH 5.4 by the sitting-drop variant of the vapour-diffusion method at 293 K in the presence of 34% polyethylene glycol monomethyl ether 2000, 0.1 M ammonium acetate and 0.2 M ammonium sulfate.
Microseeding appeared to be crucial for obtaining satisfactory crystals. Initial crystals of NOX were crushed and diluted by vortex in different amounts of 30% polyethylene glycol monomethyl ether 2000, 0.1 M sodium acetate and 0.2 M ammonium sulfate pH 5.4; these served as seeding stocks. 0.5 µl of the different seeding stocks were added to a drop which consisted of 5 µl NOX (2.1 mg ml−1) and 1 µl reservoir equilibrated against 300 µl reservoir solution. The optimized reservoir consisted of 34% polyethylene glycol monomethyl ether 2000, 15% glycerol, 0.1 M sodium acetate and 0.2 M ammonium sulfate pH 5.4. As the quality and the size of the crystals depended on the quality of the seed crystals, several cycles of seeding were performed.
Crystals grew to dimensions of 0.2 × 0.2 × 0.1 mm in about one month. Whereas initial crystallization led to irregular crystals (which grew within approximately 5 d), several cycles of seeding led to good crystals of tetragonal shape.
2.3. Diffraction data collection and analysis
A native data set was collected on an RU-200 rotating copper-anode generator (MSC) operating at 45 kV and 100 mA with a MAR 345 detector at 100 K. The crystal-to-detector distance was 230 mm and the oscillation range per image was 1°. The crystals were mounted in a nylon loop and flash-frozen in a nitrogen stream at 100 K (Oxford Cryosystems Cryostream) for data collection. The data set was integrated and scaled with DENZO and SCALEPACK (Otwinowski & Minor, 1997 ▶).
Self-rotation calculations were performed to search for non-crystallographic symmetry (NCS) using the fast rotation-function algorithm (Crowther, 1972 ▶) of the program GLRF (Tong & Rossmann, 1997 ▶). A native Patterson function was calculated using the program FFT from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
Several cycles of microseeding using seeds of native NOX were crucial for the growth of well ordered crystals with good diffraction qualities (Fig. 1 ▶). The crystals grew to their final size in about one month. These results show that NOX from L. brevis is very stable. Indeed, some water-forming NADH oxidases require additives for stabilization, such as NOX from L. sanfranciscensis which needs DTT (Riebel et al., 2002 ▶) or the enzymes from Leuconostoc mesenteroides and Serpulina hyodysenteria which require l-cysteine (Koike et al., 1985 ▶; Stanton & Jensen, 1993 ▶). DTT was also essential for the enzyme stabilization of H2O2-forming NADH oxidase from Archaeoglobus fulgidus (Reed et al., 2001 ▶).
Figure 1.

Crystals from L. brevis NADH oxidase with dimensions of approximately 0.2 × 0.2 × 0.1 mm.
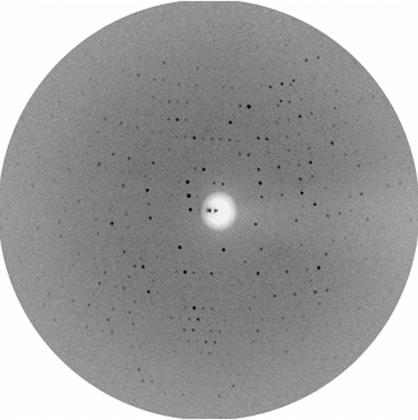
The X-ray diffraction data (Fig. 2 ▶) showed tetragonal Laue symmetry. Scaling and inspection of systematic absences revealed that the crystals belong to space group P43212 or its enantiomorph P41212. The symmetry R value is 14.7% (Table 1 ▶). The unit-cell parameters are a = b = 141.85, c = 247.62 Å. Assuming four NOX monomers per asymmetric unit, this lattice leads to a V M value of 3.2 Å3 Da−1, corresponding to a solvent content of 61.1% of the crystallographic unit cell (Matthews, 1968 ▶).
Figure 2.
X-ray diffraction pattern of a tetragonal NOX crystal.
Table 1. Statistics of an X-ray data set from native NOX.
Values in parentheses are for the last resolution shell.
| Wavelength (Å) | 1.54178 |
| Resolution range (Å) | 20–4.0 (4.14–4.0) |
| Space group | P41/3212 |
| No. of measured reflections | 75928 |
| No. of unique reflections | 21788 |
| Unit-cell volume (Å3) | 4.98 × 106 |
| Completeness (%) | 99.3 (99.7) |
| I/σ(I) | 8.0 (4.3) |
| Rsym† (%) | 14.7 (25.1) |
R
sym = 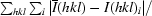
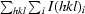 .
.
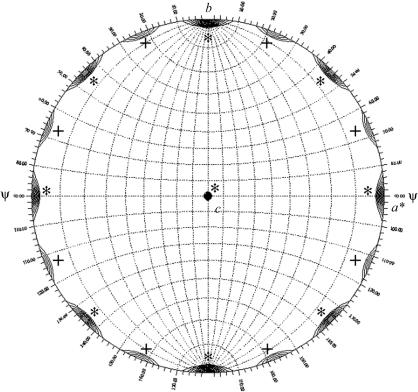
A 180° section of a self-rotation function (Fig. 3 ▶) shows a pattern of peaks in the ab plane, indicating crystallographic and non-crystallographic twofold axes perpendicular to the crystal c axis. The non-crystallographic peaks are consistent with a 222-symmetry tetramer with one of the molecular dyads parallel to the crystallographic c axis. Such an arrangement should lead to a second tetramer in the unit cell with the same orientation and hence to a pseudo-origin peak in a native Patterson function. We checked this but did not find such a peak. The reason may be that the molecular dyad is not exactly parallel to the crystallographic c axis. These and other details will be revealed by the structure determination, which is currently in progress.
Figure 3.
Self-rotation function with κ = 180°. The self-rotation function was calculated with GLRF (Tong & Rossmann, 1997 ▶) using diffraction data between 15 and 4 Å. Contour lines are drawn starting at 5 r.m.s. (root mean square) deviations above the mean in intervals of 0.2 r.m.s. deviations. Crystallographic peaks are marked with a star (*) and non-crystallographic peaks with a plus (+).
Acknowledgments
Part of this work (MK and WH) was supported by the ‘Deutsche Bundesstiftung Umwelt (DBU)’ (Förderschwerpunkt Biotechnologie, ICBio, Az 13094).
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Crowther, R. A. (1972). Int. Sci. Rev. Ser.13, 173–178.
- Higuchi, M., Yamamoto, Y., Poole, L. B., Shimada, M., Sato, Y., Takahashi, N. & Kamio, Y. (1999). J. Bacteriol.181, 5940–5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel, W., Kuzu, M. & Geueke, B. (2005). In the press.
- Hummel, W. & Riebel, B. (2003). Biotechnol. Lett.24, 51–54. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411. [Google Scholar]
- Kawasaki, S., Ishikura, J., Chiba, D., Nishino, T. & Niimura, Y. (2004). Arch. Microbiol.181, 324–330. [DOI] [PubMed] [Google Scholar]
- Koike, K., Kobayashi, T., Ito, S. & Saitoh, M. (1985). J. Biochem.97, 1279–1288. [DOI] [PubMed] [Google Scholar]
- Marty-Teysset, C., de la Torre, F. & Garel, J. (2000). Appl. Environ. Microbiol.66, 262–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Park, H. J., Kreutzer, R., Reiser, C. O. A. & Sprinzl, M. (1992). Eur. J. Biochem.205, 875–879. [DOI] [PubMed] [Google Scholar]
- Parsonage, D. & Claiborne, A. (1995). Biochemistry, 34, 435–441. [DOI] [PubMed] [Google Scholar]
- Peterson, S. N., Hu, P. C., Bott, K. E. & Hutchinson, C. A. (1993). J. Bacteriol.175, 7918–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, D. W., Millstein, J. & Hartzell, P. L. (2001). J. Bacteriol.183, 7007–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riebel, B. R., Gibbs, P. R., Wellborn, W. B. & Bommarius, A. S. (2002). Adv. Synth. Catal.344, 1156–1168.
- Sakamoto, M. & Komagata, K. (1996). J. Ferment. Bioeng.82, 210–216.
- Sakamoto, M., Uchimura, T. & Komagata, K. (1996). J. Ferment. Bioeng.82, 531–537.
- Schmidt, H. L., Stocklein, W., Danzer, J., Kirch, P. & Limbach, B. (1986). Eur. J. Biochem.156, 149–155. [DOI] [PubMed] [Google Scholar]
- Stanton, T. B. & Jensen, N. S. (1993). J. Bacteriol.175, 2980–2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, L. & Rossmann, M. (1997). Methods Enzymol.276, 594–611. [PubMed] [Google Scholar]
- Ward, D. E., Donnelly, C. J., Mullendore, M. E., van der Oost, J., de Vos, W. M. & Crane, E. J. (2001). Eur. J. Biochem.268, 5816–5823. [DOI] [PubMed] [Google Scholar]