The hybrid moelcule of calmodulin and calmodulin-binding domain of olfactory nucleotide-gated ion-channel peptide (CaM-OLFp) was crystallized and preliminary analyzed using X-ray diffaction.
Keywords: calmodulin-OLFp hybrid molecule, calmodulin-binding domains
Abstract
A hybrid molecule consisting of calmodulin (CaM) and the CaM-binding domain of olfactory nucleotide-gated ion-channel peptide (CaM-OLFp) was purified and crystallized by the hanging-drop vapour-diffusion method at 298 K. The crystals diffracted to a maximum resolution of 1.85 Å at cryogenic temperature (100 K) using X-rays from a rotating anode (Cu, wavelength 1.54 Å). The crystal belongs to the monoclinic space group C2, with unit-cell parameters a = 64.76, b = 36.23, c = 70.96 Å, α = γ = 90, β = 109.4°. Analysis of the packing density shows that the asymmetric unit contains one CaM-OLFp hybrid molecule with a solvent content of 36.42%.
1. Introduction
Calmodulin (CaM) is a highly conserved 17 kDa eukaryotic Ca2+-binding protein. In response to a Ca2+ signal, CaM interacts with and regulates various proteins, including calmodulin-dependent protein kinases, calcineurin, skeletal and smooth-muscle myosin light-chain kinases, nitric oxide synthase, cyclic nucleotide-gated ion channels, small conductance Ca2+-activated K+ channels etc. (for a review, see Hoeflich & Ikura, 2002 ▶). The three-dimensional structures of Ca2+-bound CaMs from many species have been determined (Babu et al., 1988 ▶; Taylor et al., 1991 ▶; Rao et al., 1993 ▶; Wilson & Brünger, 2000 ▶). The molecule comprises four Ca2+-binding EF-hands; the first two EF-hands form a globular N-terminal domain that is separated by a short flexible linker from a highly homologous C-terminal domain consisting of EF-hands 3 and 4. When four Ca2+ ions bind to CaM, CaM undergoes a conformational rearrangement and subsequently exposes its two hydrophobic patches for target binding (Zhang et al., 1995 ▶; Kuboniwa et al., 1995 ▶; Persechini et al., 1996 ▶). No high-resolution structure of a complex of CaM and its full-length target enzyme is available to date. Therefore, three-dimensional structures of complexes of CaM and various CaM-binding domains of its target enzymes have been used as mimetics in the past decade (Ikura et al., 1992 ▶; Meador et al., 1992 ▶ 1993 ▶; Porumb et al., 1994 ▶; Elshorst et al., 1999 ▶; Osawa et al., 1999 ▶; Kurokawa et al., 2001 ▶; Schumacher et al., 2001 ▶; Drum et al., 2002 ▶; Yap et al., 2003 ▶). Based on the solved structures, three classes of complexes of CaM and CaM-binding domains with stoichiometric ratios (CaM:CaM-binding peptide) of 1:1, 1:2 and 2:2 have been categorized (Hoeflich & Ikura, 2002 ▶). Among the solved structures in the 1:1 class, three basic Ca2+-dependent CaM-binding types, 1–10, 1–14 and 1–16, have been identified and named according to the spacing between the two anchoring hydrophobic residues in the target peptide, which forms an α-helical structure (Rhoads & Friedberg, 1997 ▶; Yap et al., 2000 ▶; Ikura et al., 2002 ▶). The widely adopted structural model in the 1–14 and 1–10 binding types is one in which the α-helical target peptide lies in a hydrophobic channel consisting of the two domains of CaM. The predominant interactions included mostly hydrophobic interactions between the N- and C-terminal portions of the binding peptide and the hydrophobic pockets of the C- and N-terminal globular domains of CaM, respectively. The 1–16 type of binding was found in the structure of CaM in complex with a CaM-dependent protein kinase kinase (CaMKK) peptide (Osawa et al., 1999 ▶); this recognition mode involves two anchor residues spaced by 14 residues. Interestingly, the orientation of the α-helix of the CaMKK peptide with respect to the two domains of CaM is the opposite of that of the peptides in the 1–14 and 1–10 types. In addition to the 1:1 binding class, complex structures involving one CaM and two peptides are classified into the 1:2 CaM-binding class (Drum et al., 2002 ▶; Yap et al., 2003 ▶). The complex of CaM and the CaM-binding domain of rat Ca2+-activated K+ channel is classified as a 2:2 CaM-binding class and involves two CaM molecules and two rat Ca2+-activated K+ channel peptides (Schumacher et al., 2001 ▶).
Olfactory cyclic nucleotide-gated ion channels (OLF channels) mediate olfactory transduction in olfactory receptor neurons (Chen & Yau, 1994 ▶; Varnum & Zagotta, 1997 ▶). They control the flow of Na+ and Ca2+ into these cells in response to signal-induced changes in the intracellular levels of cyclic nucleotides (Nilius & Droogmans, 2001 ▶). Upon binding cyclic nucleotides to its cytoplasmic C-terminal region, the OLF channel undergoes allosteric rearrangement of its conformation and thus promotes the opening of a channel pore. The opening of OLF leads to a rise in the cytosolic concentration of Ca2+. Upon binding to Ca2+, CaM disrupts the open conformation by binding to the CaM-binding domain in the N-terminal region and triggers the closure mechanism (Orsale et al., 2003 ▶). The CaM-binding domain of the OLF channel (OLFp; Arg62–Arg87, 26 residues) has the same spacing of hydrophobic residues as the M13 peptide and can be classified as a 1–14 CaM-binding motif (Porumb et al., 1994 ▶). The most interesting feature of the CaM-binding domain of the OLF channel is that it contains ‘centre-symmetrical’ segments (RIV and VIR) in its sequence between the two anchoring residues. It is important to know how OLFp interacts with the two homologous domains of CaM. Hence, we are characterizing the structure of CaM and the CaM-binding domain of the OLF channel. This study may serve as a structural model for the regulatory action of CaM on the OLF channel and will also complement the full picture on the recognition network of CaM.
Here, we report the crystallization and preliminary X-ray diffraction analysis of a hybrid molecule which contains the sequences of CaM and the CaM-binding domain of the OLF channel. These data will serve as a start towards structure determination.
2. Protein expression and purification
The gene coding for the CaM-OLFp hybrid molecule was synthesized by a series of PCR reactions using the chicken CaM gene as a template. The 5′-end primer contained an NdeI restriction-enzyme cleavage site and the 3′-end primers contained sequences coding for OLFp, a peptide linker, a stop condon and an XhoI restriction-enzyme cleavage site. The PCR-amplified CaM-OLFp hybrid gene was then subcloned into a modified pET29 expression vector (Studier et al., 1990 ▶). The construct was verified by DNA sequencing of the entire coding region and the cloning sites. The construct of CaM-OLFp hybrid protein comprises the full-length CaM followed by a pentapeptide linker (GGGGS) and residues 62–87 of the OLF channel (OLFp, 62RRGRGGFQRIVRLVGVIRDWANKNFR87) as shown in Fig. 1 ▶. The plasmid coding for the CaM-OLFp hybrid sequence was transformed into Escherichia coli strain BL21 (DE3). Cells were cultured to an OD600 of 0.8 in LB media containing 30 µg ml−1 kanamycin at 310 K and expression was induced using 0.4 mM isopropyl-β-d-thiogalactoside (IPTG). The harvested cells were resuspended in 25 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5 mM PMSF and disrupted by sonication. After the cell debris had been pelleted by centrifugation, the supernatant was loaded onto a Q-Sepharose ion-exchange column equilibrated with 25 mM Tris–HCl pH 8.0. After washing with 0.2 M NaCl, the bound proteins were eluted with a linear gradient of NaCl (0.2–0.5 M NaCl). The fractions containing CaM-OLFp hybrid proteins were concentrated and then applied onto a size-exclusion column (Hiload 16/60 Superdex 75, Pharmacia Inc.). The fractions eluted from each chromatographic step were subjected to SDS–PAGE to check the purity. The portions containing pure CaM-OLFp hybrid protein were pooled, extensively dialyzed against deionized water and then lyophilized. The final yield was 10 mg of pure CaM-OLFp hybrid protein from 1 l culture. A CaM-OLFp hybrid protein stock solution (20 mg ml−1) in 25 mM Tris–HCl, 5 mM CaCl2 pH 8.0 was prepared for crystallization trials. The concentration of CaM-OLFp hybrid protein stock solution was determined spectrophotometrically using an extinction coefficient of 8250 M −1 cm−1 at 280 nm.
Figure 1.
Schematic diagram of the construct of the CaM-OLFp hybrid protein. The CaM-OLFp hybrid protein comprises full-length CaM located at the N-terminus, OLFp (residues 62–87 of the OLF channel) positioned at the C-terminus and a peptapeptide linker (GGGGS) placed between the CaM and OLFp.
3. Crystallization and X-ray diffraction data collection
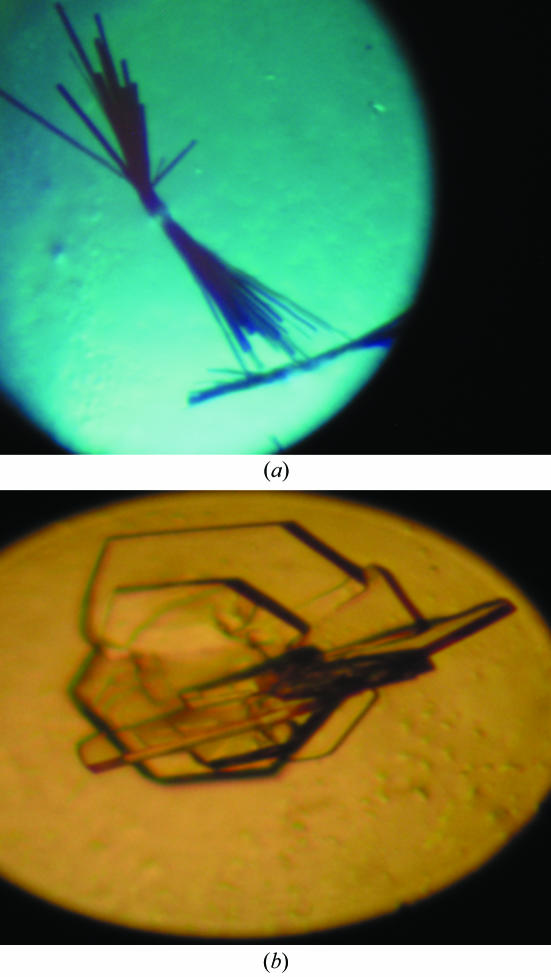
CaM-OLFp hybrid protein was crystallized using the hanging-drop vapour-diffusion method at room temperature. A modified crystal-screening protocol adopted from the standard Crystal Screen kit (Hampton Research, USA) was applied for the initial crystal screening. Needle-like crystals appeared using 30%(v/v) PEG 8000 as precipitating agent and 100 mM sodium acetate pH 4.8, 5 mM CaCl2 and 0.1% sodium azide within 20 d (Fig. 2 ▶ a). This condition was further optimized by varying the precipitant concentration, salt concentrations and the pH value of the buffer. After optimization from the initial conditions, two types of plate-shaped crystals were obtained using two sets of conditions. A thin plate-shaped crystal with dimensions 0.15 × 0.2 × 0.05 mm was obtained within 14 d using the conditions 32%(v/v) PEG 8000, 10 mM CaCl2 and 100 mM sodium acetate pH 4.6. A thick plate-shaped crystal with dimensions 0.6 × 0.5 × 0.05 mm was obtained within a week using the conditions 20%(v/v) PEG 4000, 5 mM CaCl2 and 50 mM sodium cacodyate at pH 6.0 (Fig. 2 ▶ b). The thick plate-shaped crystal was used for further diffraction analysis.
Figure 2.
Two crystal forms of CaM-OLFp hybrid molecules. (a) Rod-shaped crystals obtained from 30%(v/v) PEG 8000, 100 mM sodium acetate pH 4.8, 5 mM CaCl2 and 0.1% sodium azide with dimensions of 0.15 × 0.2 × 0.05 mm. (b) The thick plate-shaped crystals grown in 0%(v/v) PEG 4000, 50 mM sodium cacodyate pH 6.0 and 5 mM CaCl2 with dimensions of 0.6 × 0.5 × 0.05 mm. The diffraction data were collected using the crystal form in (b).
The protein crystal was mounted in a 0.5 mm nylon-fibre loop and socked in 15% glycerol cryoprotectant. A diffraction data set was collected to 1.85 Å resolution at 100 K using a rotating copper tube (R-AXIS RU-300, Rigaku, wavelength 1.54 Å) operating at 50 kV and 100 mA equipped with an image-plate detector (R-AXIS IV++ image-plate system, Rigaku). The distance from the crystal to the image plate was 150 mm, the exposure time was 10 min and the oscillation angle was 1.0°. A total of 360° of data were collected. All diffraction data collection was performed at the Department of Medical Research and Education, Veterans General Hospital Taipei.
4. Preliminary crystallographic analysis
Diffraction data were processed using the DENZO and SCALEPACK programs (Otwinowski & Minor, 1997 ▶). Systematic absence analysis suggests that the space group of the CaM-OLFp hybrid molecule crystal belongs to the C-centred monoclinic lattices (C2), with unit-cell parameters a = 64.76, b = 36.23, c = 70.96 Å, α = γ = 90°, β = 109.4°. There is one monomer in the crystallographic asymmetric unit, with a V M value of 1.94 Å3 Da−1. The calculated solvent content is 36.52% by volume (Matthews, 1968 ▶). Since the data analysis revealed that there were some redundant data, only 270 diffraction frames were used for further structure determination. The statistics for the data collected using 270/360 frames are listed in Table 1 ▶. A total of 187 520 measured reflections were merged into 57 654 independent reflections with 13 176 unique reflections. The completeness is 98.9%, with an R merge value of 6.9%. For the highest shell (1.85–1.88 Å), the completeness fell to 88.7% with an R merge of 42.2%. The phase was determined by the molecular-replacement method using the program AMoRe from the CCP4 package (Navaza, 1994 ▶) and the CaM–R20 peptide complex structure as a model (PDB code http://www.rcsb.org/pdb/cgi/explore.cgi?pdbId=1qtx). Model building and refinement are under way.
Table 1. Data-collection and processing statistics.
Values in parentheses refer to the highest resolution shell (1.85–1.88 Å).
| Crystal system | Monoclinic |
| Space group | C2 |
| Unit-cell parameters (Å, °) | a = 64.76, b = 36.23, c = 70.96, α = γ = 90, β = 109.4 |
| Resolution range (Å) | 50.0–1.85 |
| Independent reflections | 57654 |
| Unique reflections | 13176 |
| Completeness (%) | 98.9 (88.7) |
| Rmerge† (%) | 6.9 (42.2) |
| I/σ(I) | 11.0 (2.9) |
| VM (Å3 Da−1) | 1.94 |
| Vsolvent (%) | 36.52 |
R
merge = 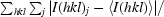
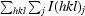 , where I(hkl)j is the jth measurement of the intensity of reflection hkl and 〈I(hkl)j〉 is the mean intensity of reflection hkl.
, where I(hkl)j is the jth measurement of the intensity of reflection hkl and 〈I(hkl)j〉 is the mean intensity of reflection hkl.
Acknowledgments
This work was supported by research grants from the National Science Council, Taiwan, NSC-93-2113-M-259-012 (to CLC) and NSC89-2113-M-320-002 and NSC89-2113-M-320-005-04 (to YCC). All diffraction experiments were performed on a Rigaku diffractometer in Veterans General Hospital Taipei, Taiwan.
References
- Babu, Y. S., Bugg, C. E. & Cook, W. J. (1988). J. Mol. Biol.204, 191–204. [DOI] [PubMed] [Google Scholar]
- Chen, T. Y. & Yau, K. W. (1994). Nature (London), 368, 545–548. [DOI] [PubMed] [Google Scholar]
- Drum, C. L., Yan, S. Z., Bard, J., Shen, Y. Q., Lu, D., Soelaiman, S., Grabarek, Z., Bohm, A. & Tang, W. J. (2002). Nature (London), 415, 396–402. [DOI] [PubMed] [Google Scholar]
- Elshorst, B., Hennig, M., Forsterling, H., Diener, A., Maurer, M., Schulte, P., Schwalbe, H., Griesinger, C., Krebs, J., Schmid, H., Vorherr, T. & Carafoli, E. (1999). Biochemistry, 38, 12320–12332. [DOI] [PubMed] [Google Scholar]
- Hoeflich, K. P. & Ikura, M. (2002). Cell, 108, 739–742. [DOI] [PubMed] [Google Scholar]
- Ikura, M., Clore, G. M., Gronenborn, A. M., Zhu, G., Klee, C. B. & Bax, A. (1992). Science, 256, 632–638. [DOI] [PubMed] [Google Scholar]
- Ikura, M., Osawa, M. & Ames, J. B. (2002). Bioessays, 24, 625–636. [DOI] [PubMed] [Google Scholar]
- Kuboniwa, H., Tjandra, N., Grzesiek, S., Ren, H., Klee, C. B. & Bax, A. (1995). Nature Struct. Biol.2, 768–776. [DOI] [PubMed] [Google Scholar]
- Kurokawa, H., Osawa, M., Kurihara, H., Katayama, N., Tokumitsu, H., Swindells, M. B., Kainosho, M. & Ikura, M. (2001). J. Mol. Biol.312, 59–68. [DOI] [PubMed] [Google Scholar]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Meador, W. E., Means, A. R. & Quiocho, F. A. (1992). Science, 257, 1251–1255. [DOI] [PubMed] [Google Scholar]
- Meador, W. E., Means, A. R. & Quiocho, F. A. (1993). Science, 262, 1718–1721. [DOI] [PubMed] [Google Scholar]
- Navaza, J. (1994). Acta Cryst. A50, 157–163. [Google Scholar]
- Nilius, B. & Droogmans, G. (2001). Physiol. Rev.81, 1415–1459. [DOI] [PubMed] [Google Scholar]
- Orsale, M., Melino, S., Contessa, G. M. & Torre, V. (2003). FEBS Lett.548, 11–16. [DOI] [PubMed] [Google Scholar]
- Osawa, M., Tokumitsu, H., Swindells, M. B., Kurihara, H., Orita, M., Shibanuma, T., Furuya, T. & Ikura, M. (1999). Nature Struct. Biol.6, 819–824. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol.276, 207–326. [DOI] [PubMed]
- Persechini, A., Gansz, K. J. & Paresi, R. J. (1996). Biochemistry, 35, 224–228. [DOI] [PubMed] [Google Scholar]
- Porumb, T., Yau, P., Harvey, T. S. & Ikura, M. (1994). Protein Eng.7, 109–115. [DOI] [PubMed] [Google Scholar]
- Rao, S. T., Wu, S., Satyshur, K. A., Ling, K. Y., Kung, C. & Sundaralingam, M. (1993). Protein Sci.2, 436–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads, A. R. & Friedberg, F. (1997). FASEB J.11, 331–340. [DOI] [PubMed] [Google Scholar]
- Schumacher, M. A., Rivard, A. F., Bachinger, H. P. & Adelman, J. P. (2001). Nature (London), 410, 1120–1124. [DOI] [PubMed] [Google Scholar]
- Studier, F. W., Rosenberg, A. H., Dunn, J. J. & Dubendorff, J. W. (1990). Methods Enzymol.185, 60–89. [DOI] [PubMed] [Google Scholar]
- Taylor, D. A., Sack, J. S., Maune, J. F., Beckingham, K. & Quiocho, F. A. (1991). J. Biol. Chem.266, 21375–21380. [DOI] [PubMed] [Google Scholar]
- Varnum, M. D. & Zagotta, W. N. (1997). Science, 278, 110–113. [DOI] [PubMed] [Google Scholar]
- Wilson, M. A. & Brünger, A. T. (2000). J. Mol. Biol.301, 1237–1256. [DOI] [PubMed] [Google Scholar]
- Yap, K. L., Kim, J., Truong, K., Sherman, M., Yuan, T. & Ikura, M. (2000). J. Struct. Funct. Genomics, 1, 8–14. [DOI] [PubMed] [Google Scholar]
- Yap, K. L., Yuan, T., Mal, T. K., Vogel, H. J. & Ikura, M. (2003). J. Mol. Biol.328, 193–204. [DOI] [PubMed] [Google Scholar]
- Zhang, M., Tanaka, T. & Ikura, M. (1995). Nature Struct. Biol.2, 758–767. [DOI] [PubMed] [Google Scholar]