A plant- and prokaryote-conserved domain (PPC) has been crystallized. The crystal diffracted to 1.7 Å resolution and belonged to space group P6322.
Keywords: Pyrococcus horikoshii, PPC domain, AHL1
Abstract
A plant- and prokaryote-conserved domain (PPC) has previously been found in AT-hook motif nuclear localized protein 1 (AHL1) localized in the nuclear matrix of Arabidopsis thaliana (AtAHL1). AtAHL1 has a DNA-binding function. Mutation analyses of AtAHL1 has previously revealed that the hydrophobic region of the PPC domain is essential for its nuclear localization. In this study, the PPC of the hyperthermophilic archaebacterium Pyrococcus horikoshii (PhPPC) was crystallized using the hanging-drop vapour-diffusion method. The crystals belonged to the hexagonal space group P6322, with unit-cell parameters a = b = 53.69, c = 159.2 Å. Data were obtained at 100 K, with diffraction being observed to a resolution of 1.7 Å. A complete data set from crystals of the SeMet-substituted protein was also obtained.
1. Introduction
AT-hook motif nuclear localized protein 1 (AHL1), which is localized in the nuclear matrix, has previously been identified in Arabidopsis thaliana using a random GFP–cDNA fusion method (Fujimoto et al., 2004 ▶). A. thaliana AHL1 (AtAHL1) is composed of a domain containing the AT-hook motif with DNA-binding ability and a PPC domain (plant- and prokaryote-conserved domain; Fujimoto et al., 2004 ▶). Phylogenetic analysis revealed that the PPC domain is conserved in AHL1 homologues and also in bacteria and archaea; however, yeasts and animals do not possess a protein with this domain. In addition, mutation analyses of AtAHL1 have indicated that the hydrophobic region of the PPC domain is essential for its nuclear localization (Fujimoto et al., 2004 ▶). However, the other functions of this domain remain unknown.
A. thaliana is highly suited to genome analysis and its genome database contains a large amount of verified data on the functional and structural properties of gene products from this organism. However, in a preliminary study we found that overexpression of AtPPC (gi:23506149) as a soluble form in Escherichia coli was rather difficult. Recently, many structural genome projects have targeted the analysis of the three-dimensional structures of proteins not only from eukaryotes, such as mice and humans, but also prokaryotes, such as Pyrococcus horikoshii OT3 and Thermus thermophilus HB8, using high-throughput determination methods. Prokaryotic PPC protein possesses a nearly equal sequence length to that of AtPPC. P. horikoshii PPC protein (PhPPC; gi:3257212) consists of 143 amino acids with a molecular weight of 16 093 Da. P. horikoshii is a hyperthermophilic archaeon that grows at temperatures ranging from 361 to 377 K and optimally at 371 K (Kawarabayasi et al., 1998 ▶). Proteins produced by this hyperthermophilic archaeon are highly stable against heat and chemical denaturants and are thus suitable for structural and functional analysis over a wide temperature range (Miyazono et al., 2004 ▶).
According to recent advances in structural proteomics, the three-dimensional structure of PhPPC at high resolution would provide essential information on the functional aspects of this protein. Here, we report the overexpression, purification, crystallization and X-ray analysis of the native and selenomethionyl (SeMet) PhPPC proteins.
2. Materials and methods
2.1. Expression and purification
The PPC gene was amplified by polymerase chain reaction (PCR) from the genomic DNA of P. horikoshii OT3. PCR was performed using the following 5′ and 3′ primers: 5′-GGAATTGCATATGGTGACTGGGATGTTC-3′ and 5′-GGAATTCTTAAAGTTTTAATTCTTCCCAAAG-3′, which were designed to contain NdeI and EcoRI sites (in bold), respectively. The PCR product was digested with NdeI and EcoRI and ligated into a pET22b(+) vector (Novagen). After sequence confirmation, the recombinant PPC was expressed in E. coli strain BL21(DE3) cells. Cells were grown in 1 l LB medium containing 50 µg ml−1 ampicillin at 310 K. When the OD660 nm of the culture reached 0.6, expression of the recombinant protein was induced by the addition of 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were then cultured at 310 K for 3 h with shaking, after which they were harvested by centrifugation at 8000g for 20 min.
The cell pellet was resuspended in 25 mM Tris–HCl pH 7.4, 2.5 mM KCl and 140 mM NaCl and disrupted by sonication. The lysate was treated at 357 K for 20 min to remove E. coli proteins. After centrifugation at 50 000g for 2 h, the supernatant was applied onto a Hi-Trap Q column (Amersham Biosciences) equilibrated with buffer A (20 mM Tris–HCl pH 8.0, 1 mM EDTA and 1 mM 2-mercaptoethanol) and proteins were eluted using a linear gradient of 0–1.0 M NaCl. The fractions containing PhPPC were dialyzed against buffer B (20 mM acetic acid pH 5.0, 1 mM EDTA and 1 mM 2-mercaptoethanol). Next, the solution was applied onto a Hi-Trap SP column (Amersham Biosciences) equilibrated with buffer B. Proteins were eluted using a linear gradient of 0–1.0 M NaCl. The eluate containing PhPPC was further purified using a HiLoad Superdex 75pg column (Amersham Biosciences) equilibrated with buffer C (25 mM Tris–HCl pH 7.4, 2.5 mM KCl and 140 mM NaCl) and eluted with the same buffer. The purified protein was concentrated with YM-10 (Millipore) to 18–20 mg ml−1 and dialyzed against 20 mM HEPES pH 7.0 with 1 mM EDTA and 1 mM 2-mercaptoethanol. The purity of the protein used for crystallization was confirmed by 15% SDS–PAGE. During labelling of the PhPPC with SeMet, E. coli strain B834 (DE3) (Novagen) was used as a host for plasmid transformation and cells were grown at 310 K in M9 media including 30 mg l−1 SeMet with IPTG to a final concentration of 0.1 mM. The SeMet-substituted PhPPC was purified using the same procedure as for wild-type protein.
2.2. Crystallization and data collection
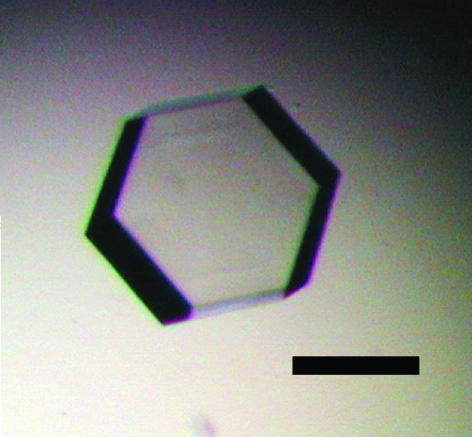
PhPPC was concentrated to 10 mg ml−1 in 20 mM HEPES pH 7.0 containing 1 mM EDTA and 1 mM 2-mercaptoethanol. Crystallization conditions were screened using Crystal Screens I and II (Hampton Research, USA) using the hanging-drop vapour-diffusion method. 1 µl protein solution was mixed in a 1:1 ratio with each of the crystallization solutions and the resultant drops were incubated at 277 K. Hexagonal plate-shaped crystals (Fig. 1 ▶) grew in the drop containing 100 mM sodium citrate pH 5.6 with 2.0 M ammonium sulfate and 0.2 M potassium sodium tartrate (Hampton Crystal Screen condition II-14) after 1 d. Subsequent refinement of the conditions gave optimal crystallization conditions involving 100 mM Tris–HCl pH 8.0 with 1.8 M ammonium sulfate and 0.2 M potassium sodium tartrate. Crystals grew to dimensions of 0.3 × 0.3 × 0.4 mm at 277 K within 5 d (Fig. 1 ▶). SeMet-substituted PhPPC was crystallized under the same conditions as those optimized for the native PhPPC. Crystals of native and SeMet-substituted PhPPC were flash-frozen using a solution comprising 100 mM Tris–HCl pH 8.0, 1.8 M ammonium sulfate, 0.2 M potassium sodium tartrate and 20%(v/v) glycerol as a cryoprotectant. X-ray diffraction data from native protein crystals were collected using Cu Kα radiation on an R-AXIS IV imaging-plate system attached to a Rigaku rotating-anode generator (MicroMax007). For SeMet-substituted crystals, X-ray diffraction data were collected at BL41XU in SPring-8 (Hyogo, Japan). Data from a native crystal and MAD data from a SeMet-substituted crystal were collected to resolutions of 1.7 and 2.6 Å, respectively. These diffraction data were integrated using MOSFLM (Leslie, 1992 ▶) and scaled using SCALA from the CCP4 program suite (Collaborative Computational Project, Number 4, 1994 ▶).
Figure 1.
A native crystal of PhPPC obtained from 100 mM Tris–HCl pH 8.0, 1.8 M ammonium sulfate and 0.2 M potassium sodium tartrate. The crystal dimensions are about 0.3 × 0.3 × 0.4 mm. Scale bar, 0.5 mm.
3. Results and discussion
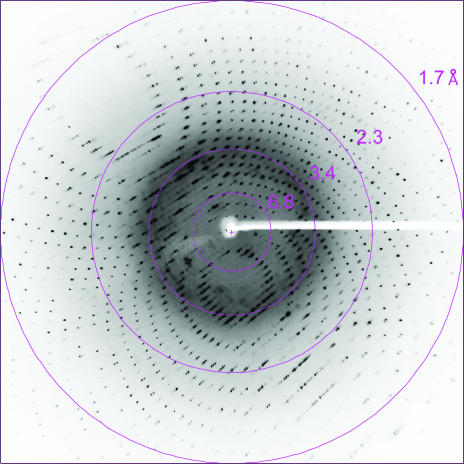
X-ray diffraction data from the crystal of native PhPPC were collected with a resolution of 1.7 Å (Fig. 2 ▶). The crystal belonged to the hexagonal space group P6322, with unit-cell parameters a = b = 53.69, c = 159.2 Å, γ = 120°. Assuming the presence of one molecule per asymmetric unit, the value of the Matthews coefficient V M and the solvent content were calculated to be 2.1 Å3 Da−1 and 41%, respectively (Matthews, 1968 ▶). SeMet-substituted PhPPC crystals diffracted to beyond 2.0 Å on a laboratory X-ray source. In order to solve the structure by the MAD method, four wavelengths, 0.9794 Å (peak), 0.9796 Å (edge), 0.9744 and 0.9843 Å (remote), were selected based on the X-ray absorption spectrum at the Se K edge. The CCD detector was placed at a distance of 200 mm and an exposure time of 4 s allowed us to obtain a 180° data set (1° frames). Data statistics are summarized in Table 1 ▶. Data were collected to 2.6 Å resolution at BL41XU in SPring-8 because the long c axis (c = 159.76 Å) of the crystals prevented the collection of high-resolution data. The data set from the SeMet-substituted PhPPC crystals was used for MAD analyses.
Figure 2.
X-ray diffraction pattern from a native crystal of PhPPC. Diffraction spots are observed to a resolution of 1.7 Å.
Table 1. Statistics of X-ray diffraction data from a SeMet PhPPC crystal.
Values in parentheses represent the outermost resolution shell.
| Edge | Peak | Remote 1 | Remote 2 | |
|---|---|---|---|---|
| Beamline | BL41XU (SPring 8) | |||
| Space group | P6322 | |||
| Wavelength (Å) | 0.9796 | 0.9794 | 0.9744 | 0.9843 |
| Unit-cell parameters (Å, °) | a = b = 54.15, c = 159.76, β = 120 | |||
| Unique reflections | 5104 | 5113 | 5164 | 5027 |
| Completeness (%) | 99.3 (99.3) | 99.3 (99.3) | 98.7 (98.7) | 98.8 (98.8) |
| Resolution (Å) | 2.54 (2.64–2.54) | 2.54 (2.64–2.54) | 2.52 (2.64–2.52) | 2.55 (2.64–2.55) |
| Rmerge (%) | 6.4 (9.9) | 6.3 (10.0) | 6.4 (9.9) | 6.4 (9.6) |
| Multiplicity | 26.2 (12.5) | 26.3 (12.5) | 15.0 (8.6) | 15.0 (8.6) |
| Average I/σ(I) | 8.2 (6.5) | 8.0 (6.2) | 7.7 (6.1) | 7.8 (6.3) |
The resolution range between 40 and 2.6 Å of the MAD data set was used for phase determination. One selenium site was found using the SOLVE program. The initial phase was modified by solvent flattening using the RESOLVE program and the phases were extended to 1.6 Å. The initial model was constructed automatically using the ARP/wARP program. On this occasion, the native data set of 1.6 Å resolution was combined with the phase output from RESOLVE and 5% of the reflections were selected for cross-validation. Refinements of the model structure of PhPPC are currently in progress.
Acknowledgments
We would like to thank Drs M. Kawamoto and N. Shimizu for their kind help during data collection at SPring-8. This study was supported in part by Special Coordination Funds from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
References
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763. [Google Scholar]
- Fujimoto, S., Matsunaga, S., Yonemura, M., Uchiyama, S., Azuma, T. & Fukui, K. (2004). Plant Mol. Biol.56, 225–239. [DOI] [PubMed] [Google Scholar]
- Kawarabayasi, Y. et al. (1998). DNA Res.5, 55–76. [DOI] [PubMed] [Google Scholar]
- Leslie, A. G. W. (1992). Crystallographic Computing 5. From Chemistry to Biology, edited by D. Moras, A. D. Podjarny & J. C. Thierry. Oxford University Press.
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed] [Google Scholar]
- Miyazono, K., Kudo, N. & Tanokura, M. (2004). Acta Cryst. D60, 1135–1136. [DOI] [PubMed] [Google Scholar]