Abstract
Aim
To investigate the role of DNA aneuploidy, particularly in patients with node negative breast cancer, in order to identify the different risk profiles within the pool of heterogeneous breast cancers.
Methods
Imprint smears from 370 breast carcinomas were Feulgen‐stained and measured by DNA image analysis. DNA aneuploidy was graded by the amount of aneuploid cells (DNA content >5c) and highly aneuploid cells (DNA content >9c) in a breast tumour population. These results were correlated to the clinical long‐term follow‐up. A statistical cut‐off value of >10 aneuploid cells (>5c) and of >1 highly aneuploid cell (>9c) was evaluated as significant for disease‐free survival (DFS) and overall survival (OS).
Results
Subgroups among patients with breast cancer with aneuploid cells below the cut‐off value showed a significantly longer DFS and OS than those with aneuploid cells above this value. Patients with node negative breast cancer with >10 aneuploid cells (>5c) and >1 highly aneuploid cell (>9c) showed an unfavourable prognosis similar to patients with node positive breast cancer with <10 aneuploid cells (>5c) and <1 highly aneuploid tumour cell (>9c) in DFS and OS.
Conclusion
Nuclear DNA content, as an objective marker of tumour aggressiveness, provides prognostic information in patients with both node negative and node positive breast cancer. Based on DNA aneuploidy, the clinically inhomogeneous group of patients with node negative breast cancer can be stratified into low‐risk and high‐risk subgroups. Therefore, DNA ploidy analysis may identify high‐risk patients with lymph node negative breast cancer who might benefit from additional adjuvant therapy.
Keywords: DNA aneuploidy; 5c‐ and 9c‐exceeding rate; DNA image cytometry; Auer classification, breast cancer; prognosis
Breast cancer is the most common malignant neoplasm in women and has an unpredictable clinical course.1 Established clinicopathological parameters currently used to assess prognosis in breast cancer often fail to characterise the clinical heterogeneity of the disease, particularly with respect to the tumour behaviour in each individual case. The presence of axillary lymph node metastases is an important criterion for adjuvant treatment of patients with breast cancer. However, up to 30% of patients with node negative breast cancer develop distant metastases after surgery, indicating a poor prognosis.2,3 Currently, the identification of subgroups in patients with node negative breast cancer who might be at a low or high risk for tumour recurrence remains difficult, despite promising attempts.4,5
In recent years, several studies have investigated potential prognostic parameters for human breast cancer in order to identify features that could be clinically useful in assessing prognosis.6,7 One of the most assessed modern prognostic markers in breast cancer is DNA ploidy.7,8,9 DNA ploidy in the nuclei of neoplastic cells measured by DNA image cytometry reflects nuclear DNA content. Neoplastic cells with irregularly increased DNA content (aneuploidy) exhibit chromosomal instability, resulting in a higher invasiveness and possibly an increased risk of metastatic spread compared to tumour cells with regular DNA content (euploidy).10,11,12,13Auer et al were the first to investigate morphometric parameters in breast cancer using DNA image cytometry.6,7,9,10,11 They classified four histogram types (Auer I–IV) according to the DNA profile of measured malignant breast cancer cells.10 In several studies, they showed that breast carcinomas with normal proliferating cell populations (Auer I and Auer II) exhibit DNA ploidy that indicates low proliferative activity. In contrast, DNA histograms (Auer III and IV) are correlated with DNA ploidy that indicates irregularly high proliferative activity.10,11
Based on these observations, we aimed to obtain prognostic information beyond clinicopathological features, and determined the impact of tumour DNA aneuploidy in relation to other established prognostic parameters, with respect to patients with lymph node negative breast cancer.
Materials and methods
Patients and tumour samples
From July 1988 to July 2000, imprint smears from 370 primary breast carcinomas of 333 female patients were sampled at the Department of Gynaecology and Obstetrics, Charité‐Campus Benjamin Franklin Berlin. All patients underwent axillary lymph node sampling to exclude distant metastases. In partnership with the Institute of Pathology, imprint smears were prepared by pressing a glass slide on the sliced breast carcinoma. The sampled material consisted of single cells or cell clusters that were considered to be a representative sample of the entire tumour cell population. Several imprints were taken from each patient to guarantee the reliability of the material by collecting cells from different parts of the breast tumour.
Excised tumours were histologically staged following the revised international TNM system criteria 200214 and were classified according to the pathological stage, histological tumour type and histological grading.15 Hormone receptor status (oestrogen and progesterone receptors) was analysed by immunocytochemistry.16,17 Tumour proliferation was evaluated by immunohistochemical staining using the Ki‐67 monoclonal antibody.18
Cytometrical DNA image measurements
Three hundred and seventy Feulgen‐stained19 breast cancer imprints were measured by quantitative DNA image analysis; this method assesses the DNA distribution patterns of tumour cell populations. DNA measurements were made as slide measurements using a microscope (Diastar/Reichert, Germany), equipped with a video‐CCD‐camera and connected to a TV‐based image analysis system (CAS 200, Becton Dickinson Co., Leiden, The Netherlands).
Each imprint smear was additionally stained by the specific Papanicolaou method20 to ensure that tumour cells were represented.
DNA measurements were made at the absorption maximum for the Feulgen stain, 546 nm, directly on the slides in areas chosen at random and scanned systematically all over the tumour. After calibration with up to 30 human lymphocytes as internal reference cells to define the normal diploid (2c) value, 100 tumour cells from the same tumour imprint were measured.
In each case, at least 10 fields per slide were selected and analysed by acquisition, digitalisation and automatic processing (fig 1). This number of fields was representative of the imprint slide and was available in all cases. Nuclei with indistinct nuclear membranes, lying close together or overlapping each other were avoided. Tumour cells were identified by two independent expert pathologists who distinguished those from normal cells by either their nuclear morphology or the additional use of phase contrast.
Figure 1 Imprint smear with microscopically typical Feulgen‐stained breast cancer cells assessed by DNA image cytometry (40×).
Cytometrical measurements were calculated automatically by measuring the nuclear integrated optical density, which represents the cytometrical equivalent of its DNA content. To compare the DNA distribution in the various tumours, DNA content was expressed in 2c units, with 2c (7.14 pg) representing the mean DNA value of the normal diploid cells of the individual specimen. To distinguish non‐diploid cells from diploid cells, an upper limit of 2.5c was set for diploid values. In accordance with international guidelines, including the updated ESACP consensus report,21,22,23 tumour cells above the 2.5c limit were taken as a measure of non‐diploidy, whereas tumour cells with DNA values above the 5c level (5c‐exceeding rate with DNA values exceeding those of proliferating diploid cells) was taken as a measure of DNA aneuploidy, and tumour cells above 9c level (9c‐exceeding rate with values exceeding the 9c level) as a measure of high DNA aneuploidy. In order to standardise DNA measurement performance and result interpretation, thresholds like 5c‐ and 9c‐exceeding rate were precisely measured in each imprint smear. In some specimens, 5c‐ and 9c‐exceeding rate values are very close to non‐tumour cells, especially in virus infected cells or cells influenced under investigation by the method. Therefore, only representative tumour type specific histograms were chosen for further histogram interpretation.
DNA histogram interpretation
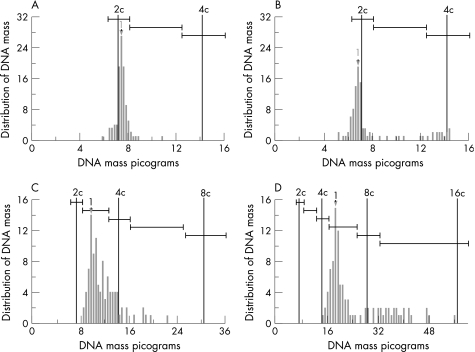
DNA histograms display the frequency distribution of DNA content values of a cell population. Histogram typing was performed according to the criteria described by Auer et al.10 According to these criteria, DNA distribution histograms were grouped into four types. Auer type I (“diploid”) is characterised by having a single distinct modal DNA value in the “diploid” or “near‐diploid” region of normal cells. Auer type II (“diploid–tetraploid”) populations show a distinct modal value in the “tetraploid” region or have two well‐defined peaks around the “diploid” and “tetraploid” regions. Auer type III (“non‐diploid”) specimens show a main peak in the “diploid” region, more than 5% in the S‐phase region, and a minor peak (less than 20%) in the “tetraploid” region. Auer type IV (“aneuploid”) populations scatter DNA values that significantly exceed the “tetraploid” region and show a very pronounced and irregular aneuploidy, with DNA amounts ranging from 2c up to values beyond 6c and 8c. This typing was based on visual criteria. Figure 2 shows representative histograms types (Auer I–IV) obtained in our patient series.
Figure 2 Histogram types according to Auer: (A) Auer I (diploid peak); (B) Auer II (tetraploid peak); (C) Auer III (triploid peak); (D) Auer IV (aneuploid peak).
Statistical analysis
Statistical analyses were undertaken using SPSS V.13.0 for Windows24 in cooperation with the Institute of Medical Informatics, Biometrics and Clinical Epidemiology at the Charité‐Campus Benjamin Franklin Berlin.
The cumulative probability of disease‐free survival (DFS) and overall survival (OS) is presented in survival curves according to Kaplan–Meier.25 The log‐rank test was used to determine the significant differences between the survival curves. In all analyses, the level of statistical significance was set at p<0.05 or p<0.01.
By using Cohen's kappa (κ) statistics, the intraobserver and interobserver agreement rates were obtained. The κ‐statistic was used to estimate agreement within categories. Conventionally, a κ‐coefficient >0.75 denotes excellent reproducibility, a κ‐value between 0.4 and 0.75 denotes moderate reproducibility and a κ‐value <0.4 denotes marginal or poor reproducibility.26,27
To determine the relative predictive strength of the prognostic variables, a Cox proportional hazards regression model28 was used with regard to DFS and OS.
Results
Patients and follow‐up
A total of 333 female patients with breast cancer were diagnosed and treated at the Department of Gynaecology and Obstetrics, Charité‐Campus Benjamin Franklin Berlin. During the follow‐up time, 6 patients died from non‐tumour related causes and 13 dropped out. Therefore, 314 of the patients were included in this long‐term follow‐up study. The median follow‐up time was 33 months (range 8–144 months).
Table 1 summarises the clinicopathological characteristics of the patients. The median age of all patients at the time of diagnosis was 57 years (range 27–89 years). Eighty three of the 314 patients were considered premenopausal and 231 were postmenopausal at the time of surgery.
Table 1 Clinicopathological parameters of patients with breast cancer (n = 314).
| Characteristics | Patients | p‐value for | ||
|---|---|---|---|---|
| % | n | DFS | OS | |
| Age | ||||
| Premenopausal | 26 | 83 | <0.040 | <0.091 |
| Postmenopausal | 74 | 231 | ||
| Tumour size (T) | <0.001 | <0.009 | ||
| pT1 | 34 | 106 | ||
| pT2 | 48 | 150 | ||
| pT3 | 6 | 20 | ||
| pT4 | 12 | 38 | ||
| Axillary node status (N) | <0.001 | <0.001 | ||
| Node negative | 49 | 155 | ||
| Node positive | 51 | 159 | ||
| Histological type | <0.001 | <0.001 | ||
| Invasive ductal | 85 | 267 | ||
| Invasive lobular | 8 | 25 | ||
| Medullar | 2 | 5 | ||
| Other types | 5 | 17 | ||
| Histological grading (G) | <0.001 | <0.001 | ||
| G1 | 12 | 38 | ||
| G2 | 45 | 140 | ||
| G3 | 35 | 136 | ||
| Hormone receptors | <0.001 | <0.001 | ||
| ER and PR positive | 65 | 203 | ||
| ER and PR negative | 22 | 70 | ||
| ER positive and PR negative | 8 | 26 | ||
| ER negative and PR positive | 5 | 15 | ||
| Auer histogram types | <0.019 | <0.323 | ||
| Auer I | 3 | 10 | ||
| Auer II | 11 | 35 | ||
| Auer III | 22 | 69 | ||
| Auer IV | 64 | 200 | ||
DFS, disease‐free survival; OS, overall survival; ER, oestrogen receptors; PR, progesterone receptors.
A total of 287 patients were treated by modified radical mastectomy and axillary lymphadenectomy as primary treatment, while 27 received conservative treatment (lumpectomy or tumourectomy) in combination with axillary lymph node dissection. In each case, at least 10 axillary lymph nodes were taken and histologically examined.
Breast cancers were staged according to the TNM stage criteria and were classified as pT1 in 34% (n = 106), pT2 in 48% (n = 150), pT3 in 6% (n = 20) and pT4 in 12% (n = 38). At the time of diagnosis none of the patients had clinically detectable distant metastases. A total of 155 patients (49%) were diagnosed as axillary node negative and 159 (51%) as node positive. Histological tumour types were grouped as invasive ductal carcinomas (85%, n = 267), invasive lobular (8%, n = 25), medullar (2%, n = 5) and other histological types (5%, n = 17). Tumour differentiation was assessed using the Bloom and Richardson grading system.40 A total of 38 patients had highly differentiated G1, 140 moderately differentiated G2 and 136 poorly differentiated G3 breast carcinomas. There were 203 hormone receptor positive (ER+ and PR+) breast carcinomas, 70 hormone receptor negative (ER− and PR−) carcinomas, 24 ER+ and PR− carcinomas and 15 ER− and PR+ carcinomas.
At the end of the follow‐up period, 224 breast cancer patients (71%) were alive without evidence of disease, 44 (14%) were alive with distant metastases, and 46 (15%) had died of their disease.
Furthermore, 81% (n = 125/155) of node negative patients were alive and recurrence‐free, 9% (n = 14/155) developed recurrences, and 10% (n = 16/155) died due to breast cancer. In node positive breast cancer, 62% (n = 99/159) were alive recurrence‐free, 19% (n = 30/159) showed recurrences, and 19% (n = 30/159) died due to the malignancy.
Reproducibility of DNA image cytometry
Independent testing of intraobserver and interobserver agreement was performed on 85 different breast cancer imprint smears (control test set) with known DNA histogram profiles. To test intraobserver agreement, each imprint was repeatedly measured by DNA image analysis and evaluated four times by one experienced researcher at different time points, resulting in an intraobserver agreement of κ = 0.77 (SD = 0.1). The interobserver agreement, tested by two independent expert pathologists, was κ = 0.52 (SD = 0.14). The level of agreement of the aneuploidy grade expressed by the κ‐coefficient was good (κ = 0.77) for intraobserver, and moderate (κ = 0.52) for interobserver evaluations.
DNA histograms and Auer classification
DNA distribution histograms from 314 patients were classified according to the method described by Auer et al.10 Figure 2 shows distinct DNA histograms of our series (Auer I–IV), which seemed to be representative for each histogram type according to Auer. Using the Auer classification, the majority of our patients (86%, n = 269) were classified in the unfavourable Auer III and Auer IV groups. However, at the end of the study, 71% (n = 224) of these patients had no distant recurrences.
Correlation between cut‐off value for aneuploidy and prognosis of patients with breast cancer
After cytometrical measurement of tumour cells with more than 5c DNA content (5c‐exceeding rate) and more than 9c DNA content (9c‐exceeding rate), statistical cut‐off values were calculated for both aneuploid >5c and highly aneuploid tumour cells >9c.
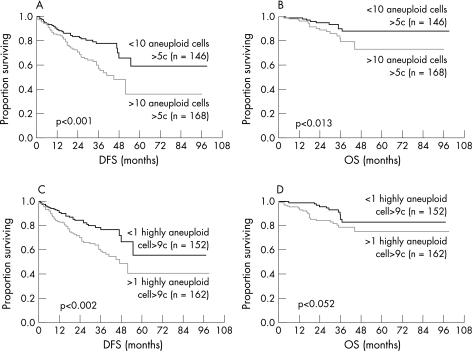
In Figure 3, Kaplan–Meier survival curves show a significant cut‐off value of more than 10 tumour cells for aneuploid >5c cells and of >1 tumour cell for highly aneuploid >9c cells. Using these statistical cut‐off values, all patients (n = 314) were divided into two groups with significant DFS and OS rates. A total of 146 patients with less than 10 aneuploid tumour cells >5c had a lower recurrence rate than 168 patients with more than 10 aneuploid tumour cells >5c (21% versus 35%, p<0.001) (fig 3A). A similar trend was observed for highly aneuploid cells >9c with less than 1 tumour cell and those with more than 1 tumour cell (20% versus 37%, p<0.0015) for DFS and OS (fig 3B). However, it is interesting that the DNA profile of patients with shorter DFS and OS differs from that of patients with longer DFS and OS in the total amount of aneuploid and highly aneuploid tumour cells.
Figure 3 (A) Disease‐free and overall survival curves (Kaplan–Meier) for patients with breast cancer, stratified according to the calculated cut‐off for aneuploid cells >5c (⩽10 aneuploid versus >10 aneuploid cells) (p<0.001 and p<0.013). (B) Disease‐free and overall survival curves stratified according to the cut‐off for highly aneuploid cells >9c (⩽1 highly aneuploid versus >1 highly aneuploid cell) (p<0.0015 and p<0.052).
Correlation between cut‐off value and axillary lymph node involvement
Table 2 shows the median number of aneuploid and highly aneuploid cells in relation to axillary lymph node involvement in 314 patients with breast cancer. In comparison, node negative patients show a lower median number of aneuploid >5c (mean 2 cells) and highly aneuploid tumour cells >9c (mean 1 cell) compared to node positive patients. In node positive patients (N1 versus N2) the mean amount of aneuploid tumour cells differs: 13 cells and 2 cells for >5c and >9c, respectively, versus 43 cells and 15 cells for >5c and >9c, respectively. Using the calculated cut‐off values, node negative and node positive patients can be divided into groups with different prognostic outcome according to the amount of aneuploid cells >5c and highly aneuploid cells >9c in DFS and OS.
Table 2 Median distribution of aneuploid cells >5c and highly aneuploid cells >9c (n = 314) in relation to axillary lymph node status.
| Axillary lymph node status (n = 314) | Median of aneuploid cells >5c | Median of highly aneuploid cells >9c |
|---|---|---|
| N0 (n = 155) | 2 | 1 |
| N1 (n = 148) | 13 | 2 |
| N2 (n = 11) | 43 | 15 |
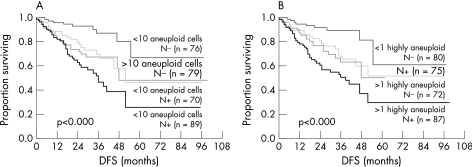
Figure 4 shows that node negative patients have a longer DFS and OS compared to node positive patients (p<0.001). Using the cut‐off values for the 5c‐ and 9c‐exceeding rate, the group of patients with node negative breast cancer (n = 155) showed two distinct prognostic subgroups: patients with a lower amount of aneuploid >5c (n = 76) and highly aneuploid tumour cells >9c (n = 80) and patients with a higher amount of aneuploid >5c (n = 79) and highly aneuploid cells >9c (n = 75). The subgroup with a lower grade of aneuploidy has a significantly longer DFS and OS compared to those with a higher grade of aneuploidy (p<0.001). Interestingly, we found that node negative patients with high grade DNA aneuploidy showed a similar poor prognosis as node positive patients with a lower rate of DNA aneuploidy (p<0.001). Thus, patients with node negative breast cancer with the additional presence of high grade DNA aneuploidy, have an unfavourable clinical outcome.
Figure 4 (A) Disease‐free survival (Kaplan–Meier) for patients with node‐negative and node‐positive breast cancer, split into two prognostic groups according to the amount of aneuploid tumour cells >5c. (B) Disease‐free survival for patients with node‐negative and node‐positive breast cancer, stratified according to the amount of highly aneuploid tumour cells >9c.
Statistical analysis
Performing the multivariate Cox regression analysis, aneuploidy (5c‐ and 9c‐exceeding rate) was included in addition to histomorphological grading, axillary lymph node status and DNA histogram classification according to Auer. Table 3 shows that histological grading and the axillary lymph node status were the strongest prognostic factors for DFS and OS, followed by the grade of aneuploidy (p<0.004 for DFS and p<0.001 for OS). The Auer classification failed to give statistically significant prognostic information (p<0.865 for DFS and p<0.965 for OS). Both histomorphological grading and DNA aneuploidy were identified as parameters adding independent prognostic information. Furthermore, these results suggest that DNA aneuploidy has significant prognostic value compared to the histogram classification according to Auer.
Table 3 Cox regression multivariate analysis of patients with breast cancer (n = 314).
| Stepwise selected variables | p‐value for | 95% CI | |
|---|---|---|---|
| DFS | OS | ||
| Histological grading | 0.0005 | 0.0094 | 1.35 to 2.62 |
| Axillary lymph node status | 0.0003 | 0.0382 | 1.31 to 2.65 |
| 5c‐exceeding rate | 0.0040 | 0.0005 | 1.00 to 1.02 |
| 9c‐exceeding rate | 0.0151 | 0.0185 | 0.95 to 1.01 |
| Auer classification | 0.8650 | 0.9649 | 0.85 to 1.03 |
DFS, disease‐free survival; OS, overall survival.
Discussion
Breast cancer is the most common malignant disease in women. In recent years, several prognostic parameters have been used as indicators of disease progression in breast cancer.10,11,29,44,45 One of the most investigated prognostic factor in breast cancer is the axillary lymph node status.2,4,5,30,31,51 Since node positive patients have a higher risk of disease progression with shorter disease‐free survival und overall survival than node negative patients, the involvement of axillary lymph nodes proved to be one important prognostic indicator.1,4,32 As a consequence, node positive breast cancer patients receive intense systemic treatment. However, distant metastases do develop in up to 30% of node negative patients, indicating a poor clinical outcome.8,33 Currently, the identification of high‐risk subgroups in node negative breast cancer remains difficult.4,5 Therefore, the recent guidelines of the St Gallen International Expert Consensus conference on the primary therapy of early breast cancer 2005,34 newly defined three risk categories, including a group merging higher risk node negative disease (low grade disease with features conferring a worse prognosis, e.g. HER2/neu overexpression35 or lymphovascular invasion36) and lower risk node positive disease (1–3 involved axillary lymph nodes but no other adverse features) into an intermediate risk group “across nodal status”. Consequently, node negative breast cancer patients are categorised as low and intermediate risk group patients, indicating that axillary lymph node involvement is a dependent prognostic factor and can be influenced by additional risk features.
In an attempt to identify those subgroups of high‐risk node negative breast cancer patients, this study investigated the role of DNA aneuploidy as a prognostic criterion for patients' individual risk of tumour progression.
Although various studies have shown a strong correlation between DNA aneuploidy and increased risk of recurrence, the prognostic significance of DNA ploidy in breast cancer is still controversial.10,12,46,47,48,49,50,52,53,54 Using the Auer classification, we investigated the correlation between DNA histograms and the clinical outcome of patients. We found that 86% (n = 269/314) of our patients were grouped into Auer histogram types III and IV category. Comparing these results to the disease‐free and overall survival, we showed that 71% of these patients (n = 191/269) remained recurrence‐free during the follow‐up, indicating that there is not a strong correlation of Auer histogram typing and clinical outcome. These findings are in agreement with other studies, e.g. Ottesen et al,8 von Rosen et al12 and Lorenzato et al.43 However, other groups, such as Fallenius et al and Caspersson et al found a significant correlation between the Auer classification and the follow‐up of patients with breast cancer.10,11 The disagreement between various reports could be generally related to the lack of standardised methodological approaches in relation to sampling procedures or interpretation of the results.37,38 However, this controversy encouraged us to look for a more precise prognostic parameter based on DNA cytometry in a large number of patients with breast cancer. In addition, to avoid technical bias, we used the international agreed recommendations for standardised image analysis measurements.21,22,23
We defined a statistically significant cut‐off value for DNA aneuploidy to discriminate between subgroups of patients with low and high recurrence risk of breast cancer. A statistical cut‐off point of more than 10 aneuploid cells >5c (5c‐exceeding rate) and of more than one high aneuploid cell >9c (9c‐exceeding rate) was significantly calculated. Using these cut‐offs for the 5c‐exceeding rate and 9c‐exceeding rate, patients could be split into two prognostic subgroups. Patients with low grade DNA aneuploidy (less than 10 aneuploid tumour cells >5c) had a lower recurrence rate than patients with high grade DNA aneuploidy (more than 10 aneuploid tumour cells >5c) (21% versus 35%). The similar trend was observed for highly aneuploid cells >9c with less than 1 tumour cell and those with more than 1 tumour cell in 20% and 37%, respectively. Furthermore, a significant number of patients with high grade DNA aneuploidy had died due to their disease compared to patients with low grade DNA aneuploidy (17% versus 12%). In both subgroups, Kaplan–Meier survival curves revealed that both the number of aneuploid cells >5c, and highly aneuploid cells >9c determined disease‐free and overall survival.
A strongly significant influence between DNA ploidy and axillary nodal status was present in relation to disease‐free and overall survival. As shown by Aubele et al,39 DNA ploidy and morphometric parameters can provide prognostic information in patients with mainly node negative breast cancer who are at a higher risk of distant recurrence. In accordance with these data, we showed that within node negative patients, the subset of patients with high aneuploidy rates had a poorer prognosis than patients with lower aneuploidy rates. We were able to show that patients with node negative breast cancer with more than 10 aneuploid cells >5c and more than one highly aneuploid tumour cell >9c have a similarly poor prognosis as patients with node positive breast cancer with less than 10 aneuploid cells >5c, and less than 1 highly aneuploid tumour cell >9c.
Take‐home messages
Nuclear DNA content, as an objective marker of tumour aggressiveness, provides prognostic information in patients with both node negative and node positive breast cancer.
Patients with breast cancer with a high number of aneuploid cells below >10 aneuploid cells (>5c) and >1 highly aneuploid cell (>9c) showed a significant longer disease‐free survival and overall survival than those with aneuploid cells above this value.
Patients with node negative breast cancer with a high number of aneuploid cells showed an unfavourable prognosis similar to patients with node positive breast cancer.
Based on DNA aneuploidy, the clinically inhomogeneous group of patients with node negative breast cancer can be stratified into low‐risk and high‐risk subgroups.
In addition to axillary lymph node involvement and histological grading, multivariate Cox analysis showed that aneuploidy (5c‐ and 9c‐exceeding rate) correlated significantly with a poorer clinical outcome.
In conclusion, DNA image cytometry is a reproducible and reliable method which supplies powerful prognostic information by measuring DNA ploidy in breast cancer.11,12,29,31,40,41,42 We could show that DNA aneuploidy is capable of defining subgroups of patients with breast cancer according to their recurrence risk and predicting the clinical outcome in both node negative and node positive breast cancer. Moreover, our data indicate that the grade of aneuploidy allows additional classifying of patients with node negative breast cancer into low‐risk and high‐risk subgroups. As a consequence, we suggest that in high‐risk node negative patients with a high aneuploidy grade, more aggressive adjuvant chemotherapy should be considered.
Acknowledgements
This study contains parts of the doctoral thesis of S Yildirim‐Assaf. The authors gratefully acknowledge the excellent technical assistance of Mrs M Kühnert. We thank Prof. Dr Dr S Nigam and L Udvarhelyi for their editorial work.
Abbreviations
DFS - disease‐free survival
OS - overall survival
Footnotes
Competing interests: None declared.
References
- 1.Feuer E J, Wun L M, Boring C C.et al The lifetime risk of developing breast cancer. J Natl Cancer Inst 199385892–897. [DOI] [PubMed] [Google Scholar]
- 2.Yuan J, Hennessy C, Givan A L.et al Predicting outcome for patients with node negative breast cancer: a comparative study of the value of flow cytometry and cell image analysis for determination of DNA ploidy. Br J Cancer 199265461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore M P, Kinne D W. Axillary lymphadenectomy: a diagnostic and therapeutic procedure. J Surg Oncol 1997662–6. [DOI] [PubMed] [Google Scholar]
- 4.McGuire W L, Clark G M. Prognostic factors and treatment decisions in axillary node‐negative breast cancer. N Engl J Med 19923261756–1761. [DOI] [PubMed] [Google Scholar]
- 5.Gasparini G, Pozza F, Harris A L. Evaluating the potential usefulness of new prognostic and predictive indicators in node‐negative breast cancer patients. J Natl Cancer Inst 1993851206–1219. [DOI] [PubMed] [Google Scholar]
- 6.Aubele M, Auer G, Falkmer U.et al Identification of a low‐risk group of stage I breast cancer patients by cytometrically assessed DNA and nuclear texture parameters. J Pathol 1995177377–384. [DOI] [PubMed] [Google Scholar]
- 7.Aubele M, Auer G, Voss A.et al Different risk groups in node negative breast cancer: prognostic value of cytophotometrically assessed DNA, morphometry and texture. Int J Cancer 1995637–12. [DOI] [PubMed] [Google Scholar]
- 8.Ottesen G L, Christensen I J, Larsen J K.et al DNA aneuploidy in early breast cancer. Br J Cancer 199572832–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burger G, Aubele M, Jutting U.et al Interactive cytometry, chance or evil of bias? Pathol Res Pract 1992188391–395. [DOI] [PubMed] [Google Scholar]
- 10.Auer G U, Caspersson T O, Wallgren A S. DNA content and survival in mammary carcinoma. Anal Quant Cytol 19802161–165. [PubMed] [Google Scholar]
- 11.Fallenius A G, Auer G U, Carstensen J M. Prognostic significance of DNA measurements in consecutive breast cancer patients. Cancer 198862331–341. [DOI] [PubMed] [Google Scholar]
- 12.von Rosen A, Rutqvist L E, Carstensen J.et al Prognostic value of nuclear DNA content in breast cancer in relation to tumour size, nodal status, and estrogen receptor content. Breast Cancer Res Treat 19891323–32. [DOI] [PubMed] [Google Scholar]
- 13.Blegen H, Will J S, Ghadimi B M.et al DNA amplifications and aneuploidy, high proliferative activity and impaired cell cycle control characterize breast carcinomas with poor prognosis. Anal Cell Pathol 200325103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singletary E S, Allred C, Ashley P.et al Revision of the American Joint Committee on cancer staging system for breast cancer. J Clin Oncol 2002203628–3636. [DOI] [PubMed] [Google Scholar]
- 15.Bloom H J, Richardson W W. Histological grading and prognosis in breast cancer; a study of 1409 cases of which 359 have been followed for 15 years. Br J Cancer 195711359–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remmele W, Stegner H E. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER‐ICA) in breast cancer tissue. Pathologe 19878138–140. [PubMed] [Google Scholar]
- 17.Pichon M F, Broet P, Magdelenat H.et al Prognostic value of steroid receptors after long‐term follow‐up of 2257 operable breast cancers. Br J Cancer 1996731545–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdes J, Lelle R J, Pickartz H.et al Growth fractions in breast cancers determined in situ with monoclonal antibody Ki‐67. J Clin Pathol39977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feulgen R, Rossenbeck H. Mikroskopisch‐chemischer Nachweis einer Nukleinsäure vom Typus der Thymonukleinsäure und die darauf beruhende elektive Färbung von Zellkernen in mikroskopischen Präparaten. Z Physiol Chem 1924135203–248. [Google Scholar]
- 20.Papanicolaou G N, Traut H F. The diagnostic value of vaginal smears in carcinoma of the uterus. Am J Obstet Gynecol 194142193–206. [PubMed] [Google Scholar]
- 21.Giroud F, Haroske G, Reith A.et al Part I: Basic considerations and recommendations for preparation, measurement and interpretation. Anal Cell Pathol 199817189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giroud F, Haroske G, Reith A.et al Part II: Specific recommendations for quality assurance. Anal Cell Pathol 199817201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haroske G, Baak J P A, Danielsen H.et al Fourth updated ESACP consensus report on diagnostic DNA image cytometry. Anal Cell Pathol 20012389–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie H H, Hadlai H, Jenkins J G.et alSPSS (Statistical Package for the Social Sciences). New York, NY: McGraw‐Hill, 1979
- 25.Kaplan E L, Meier P. Nonparametric estimation from incomplete observations. J Am Stat Assoc 195853457–481. [Google Scholar]
- 26.Brennan P, Silman A. Statistical methods for assessing observer variability in clinical measures. BMJ 19923041491–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landis J R, Koch G G. The measurement of observer agreement for categorical data. Biometrics 197733159–174. [PubMed] [Google Scholar]
- 28.Cox D R. Regression models and life tables. J R Stat Soc B 197234187–220. [Google Scholar]
- 29.Baak J P, Van Dop H, Kurver P H.et al The value of morphometry to classic prognosticators in breast cancer. Cancer 198556374–382. [DOI] [PubMed] [Google Scholar]
- 30.Aubele M, Auer G, Hofler H. Analysis of DNA and morphometry in breast carcinoma. Histochem Cell Biol 1996106241–245. [DOI] [PubMed] [Google Scholar]
- 31.Sigurdsson H, Baldetorp B, Borg A.et al Indicators of prognosis in node‐negative breast cancer. N Engl J Med 19903221045–1053. [DOI] [PubMed] [Google Scholar]
- 32.Baak J P A, van Diest P J, Voorhorst F J.et al Prospective multicenter validation of the mitotic activity index in lymph node‐negative breast cancer patients younger than 55 years. J Clin Oncol 2005235993–6001. [DOI] [PubMed] [Google Scholar]
- 33.Stanton P D, Cooke T G, Oakes S J.et al Lacl of prognostic significance of DNA ploidy and S‐phase fraction in breast cancer. Br J Cancer 199266925–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldhirsch A, Glick J H, Gelber R D, and Panel Members et al Meeting Highlights: International Expert Consensus on the Primary Therapy of Early Breast Cancer 2005. Ann Oncol 2005161569–1583. [DOI] [PubMed] [Google Scholar]
- 35.Bofin A M, Ytterhus B, Martin C.et al Detection and quantitation of HER‐2 gene amplification and protein expression in breast carcinoma. Am J Clin Pathol 2004122110–119. [DOI] [PubMed] [Google Scholar]
- 36.Schoppmann S F, Bayer G, Aumayr K.et al Prognostic value of lymphangiogenesis and lymphovascular invasion in invasive breast cancer. Ann Surg 2004240306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hedley D W, Clark G M, Cornelisse C J.et al Consensus review of the clinical utility of DNA cytometry in carcinoma of the breast. Cytometry 199314482–485. [DOI] [PubMed] [Google Scholar]
- 38.Shankey T V, Rabinovitch P S, Bagwell B.et al Guidelines for implementation of clinical DNA cytometry. Cytometry 199314472–477. [DOI] [PubMed] [Google Scholar]
- 39.Auer G, Einhorn N, Nilsson B.et al Biological malignancy grading in early‐stage ovarian carcinoma. Acta Oncol 19963593–98. [DOI] [PubMed] [Google Scholar]
- 40.Mandard A M, Denoux Y, Herlin P.et al Prognostic value of DNA cytometry in 281 premenopausal patients with lymph node negative breast carcinoma randomized in a control trial: multivariate analysis with Ki‐67 index, mitotic count, and microvessel density. Cancer 2000891748–1757. [DOI] [PubMed] [Google Scholar]
- 41.Pinto A E, Andre S, Soares J. Short‐term significance of DNA ploidy and cell proliferation in breast carcinoma: a multivariate analysis of prognostic markers in a series of 308 patients. J Clin Pathol 199952604–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beerman H, Kluin P M, Hermans J.et al Prognostic significance of DNA‐ploidy in a series of 690 primary breast cancer patients. Int J Cancer 19904534–39. [DOI] [PubMed] [Google Scholar]
- 43.Lorenzato M, Abboud P, Masure M.et al Image cytometry detection of breast cancer cells with >5c DNA content and minor DNA stemlines. Anal Quant Cytol Histol 200022199–205. [PubMed] [Google Scholar]
- 44.Casperson T O. History of the development of cytophotometry from 1935 to the present. Anal Quant Cytol Histol 198792–6. [PubMed] [Google Scholar]
- 45.Baldetorp B, Ferno M, Fallenius A.et al Image cytometric DNA analysis in human breast cancer analysis may add prognostic information in diploid cases with low S‐phase fraction by flow cytometry. Cytometry 199213577–585. [DOI] [PubMed] [Google Scholar]
- 46.Sen S. Aneuploidy and cancer. Curr Opin Oncol 20001282–88. [DOI] [PubMed] [Google Scholar]
- 47.Keyhani‐Rofagha S, O'Toole R V, Farrar W B.et al Is DNA ploidy an independent prognostic indicator in infiltrative node‐negative breast adenocarcinoma? Cancer 1990651577–1582. [DOI] [PubMed] [Google Scholar]
- 48.Magennis D P. Nuclear DNA in histological and cytological specimen: measurement and prognostic significance. Br J Biomed Sci 199754140–148. [PubMed] [Google Scholar]
- 49.Millot C, Dufer J. Clinical applications of image cytometry to human tumour analysis. Histol Histopathol 2000151185–1200. [DOI] [PubMed] [Google Scholar]
- 50.Baldetorp B, Ferno M, Bendahl P O.et al Proliferative index obtained by DNA image cytometry. Does it add prognostic information in Auer IV breast cancer? Am J Epidemiol 199820144–152. [PubMed] [Google Scholar]
- 51.Seker H, Odetayo M O, Petrovic D.et al Assessment of nodal involvement and survival analysis in breast cancer patients using image cytometric data: statistical, neural network and fuzzy approaches. Anticancer Res 200222433–438. [PubMed] [Google Scholar]
- 52.Steinbeck R G, Auer G U, Zetterberg A D. Reliability and significance of DNA measurements in interphase nuclei and division figures in histological sections. Eur J Cancer 199935787–795. [DOI] [PubMed] [Google Scholar]
- 53.Kronenwett U, Huwendiek S, Ostring C.et al Improved grading of breast adenocarcinomas based on genomic instability. Cancer Res 200464904–909. [DOI] [PubMed] [Google Scholar]
- 54.Kronenwett U, Huwendiek S, Castro J.et al Characterisation of breast fine needle aspiration biopsies by centrosome aberrations and genomic instability. Br J Cancer 200592389–389. [DOI] [PMC free article] [PubMed] [Google Scholar]