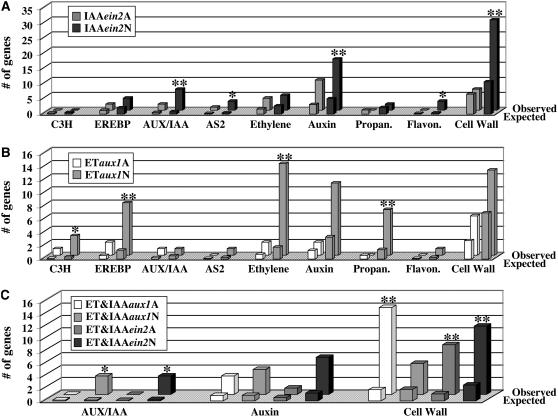
Figure 6.
Gene Functional Analysis Supports the Existence of Hormone-Specific Effects and the Effects Mediated by the Interaction between Ethylene and Auxin.
Several gene function categories were found to be significantly enriched in one of the ethylene- and/or auxin-regulated gene groups. The MapMan software and the corresponding gene function databases were used to determine the significance of the enrichment and the number of observed and expected genes in each functional group (see Methods for more details). ** and * indicate a P value < 0.0001 (with the * marking functional categories containing ≤3 genes).
(A) Comparison between the number of genes (observed versus expected) in the following functional categories of auxin-regulated genes: C3H (C3H zing finger family of transcription factors), EREBP (AP2/EREBP family of transcription factors), IAA (AUX/IAA gene family), AS2 (family of transcription factors related to AS2), ethylene (genes annotated as ethylene related or ethylene metabolism), auxin (genes annotated as auxin related or auxin metabolism), propan. (genes annotated as secondary metabolism, phenylpropanoids), flavon. (genes annotated as secondary metabolism, flavonols), and cell wall (cell wall metabolism genes).
(B) Comparison between the numbers of observed and expected genes in the same functional categories as in (A) but among ethylene-regulated genes.
(C) Comparison between the number of observed and expected genes in the following functional categories of ethylene- and auxin-regulated genes: IAA (AUX/IAA gene family), auxin (auxin-related or auxin metabolism-related genes), and cell wall (cell wall metabolism genes).