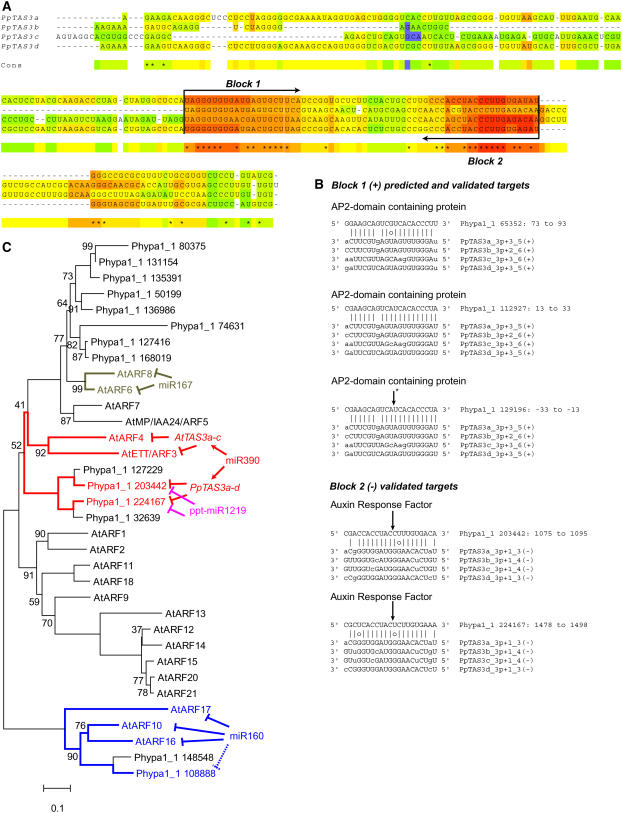
Figure 4.
Similar ARF Targets for Diverse ta-siRNAs and miRNAs.
(A) T-COFFEE alignment of the regions of Pp TAS3a-d between the two miR390 complementary sites. Two ∼21-nucleotide blocks conserved between the four paralogs are indicated.
(B) Alignments of the block 1 (+) siRNAs and the block 2 (-) RNAs with their predicted and validated targets. Lowercase letters indicate positions that cannot form either Watson-Crick or G-U pairs with the target. Arrows indicate cleavage-validated target sites. The asterisk indicates the cleavage site of the AP2 domain–containing gene Phypa1_1 129196 by Pp TAS3a-d block 1 (+)–derived siRNAs as demonstrated by Talmor-Neiman et al. (2006b).
(C) An unrooted phylogenetic reconstruction of the relationships between Arabidopsis and P. patens ARF proteins. Nodes with <100% bootstrap support are noted with the percentage of supporting replicates. ARF genes known or predicted to be under small RNA–mediated regulation are indicated. Accession numbers are given in Methods.