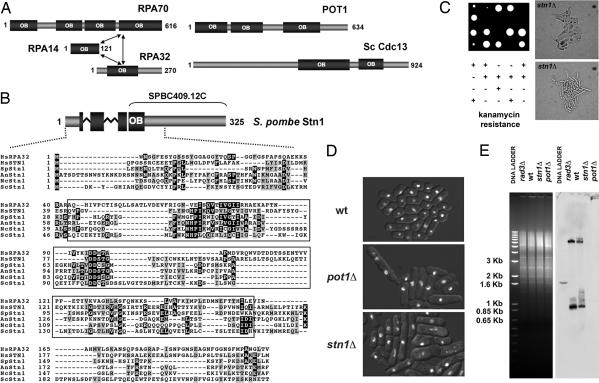
Fig. 1.
Stn1 homologs and analysis of fission yeast stn1Δ cells. (A) Schematic showing the human RPA, human POT1, and budding yeast Cdc13 OB-fold domain structure. The model for the structural organization of the RPA trimer was previously described (23). The positions of the multiple OB-fold domains in POT1 and Cdc13 are based on previously published data (16). (B) (Upper) Schematic representation of S. pombe Stn1. The protein fragment previously annotated as SPBC409.12C is shown. The N-terminal OB-fold of Stn1 is represented as a fragmented box to show the partial OB-fold present in SPBC409.12C and the one obtained by adding the N-terminal sequences obtained during our 5′ RACE analyses. Zig-zag lines indicate the position of the two introns in the 5′ region of the corresponding gene. (Lower) Multiple alignment of the N-terminal regions of the Stn1 orthologs in Hs, Homo sapiens (NP_079204); Sp, S. pombe (CAB52614); An, Aspergillus nidulans (XP_663844); Nc, Neurospora crassa (XP_960343); and Sc, S. cerevisiae (CAA98902). Alignment of human RPA32 is also provided. Residues included in the predicted OB-fold domains of the different proteins appear boxed. PSI-BLAST indicates that the sequence similarities of the OB-fold domains are highly significant [e.g., SpStn1 (3–129) vs. HsStn1 (11–158), E value = 2e−40; SpStn1 (19–160) vs. HsRPA32 (47–182), E value = 4e−21]. (C) Tetrad dissection of the heterozygous stn1Δ diploid strain (Left), and cell morphology of the stn1Δ haploid null mutants (Right). Two colonies at the ≈20-cell stage were photographed. (D) Fluorescence images of DNA stained with Hoechst 33342 were overlaid on bright-field images of the cells. Cells came from colonies formed after spores were incubated at 30°C for 3 days. The mutant colonies contained ≤2,000 cells. The mutants displayed similar phenotypes. A significant number of mutant cells are elongated or misshapen. Hoechst staining showed unevenly divided DNA in many mutant cells. (E) Loss of TAS1 telomeric-proximal DNA in stn1Δ cells. EcoRI-digested genomic DNA was subjected to electrophoresis and stained with ethidium bromide (Left) before its transfer to a membrane and hybridization to a TAS1 probe (Right). Genomic DNA from wild-type (wt), rad3Δ, and pot1Δ strains was used as controls.