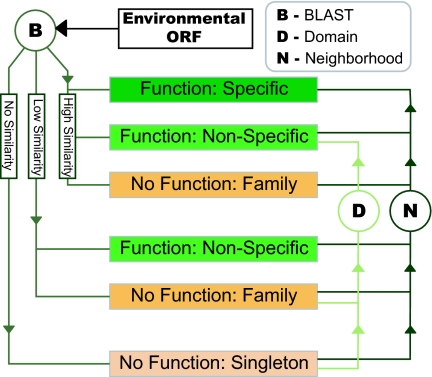
Fig. 1.
Flow chart of function prediction procedure. By using homology to genes in the KEGG, COG, and UniRef90 databases, ORFs were divided into four categories based on the level of functional annotation possible; (i) specific functional annotation: ORFs similar to genes with specific functional information; (ii) nonspecific functional annotation: ORFs similar to genes that have been characterized at a general level or low similarity; (iii) no functional annotation but member of an existing family: ORFs with homologs in one of the databases but no functional information (e.g., “conserved hypothetical”); (iv) singletons: ORFs that have no significant similarity to known sequences. ORFs containing domains from the SMART and Pfam A databases were upgraded to having nonspecific annotation where applicable. Finally genomic neighborhood methods were used to infer functional links between ORFs and upgrade the functional annotation accordingly.