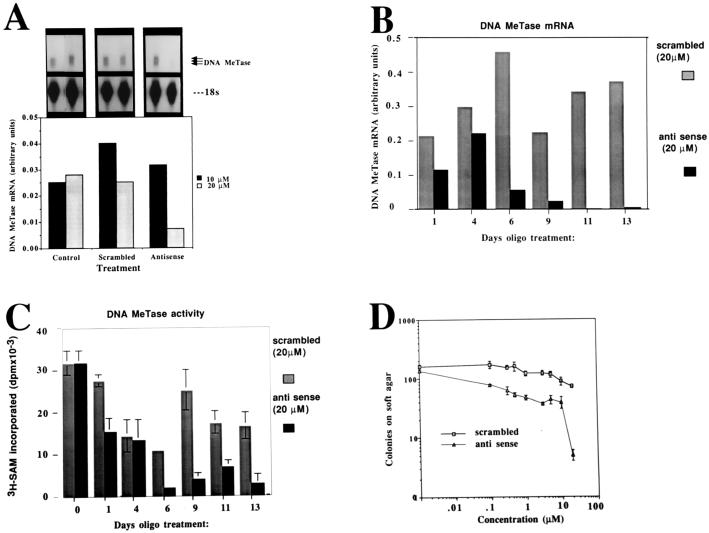
Figure 1.
DNA MeTase antisense oligodeoxynucleotides inhibit DNA MeTase mRNA, DNA MeTase activity, and anchorage independent growth ex vivo. (A) RNase protection analysis of DNA MeTase mRNA in Y1 cells treated with control scrambled and antisense oligodeoxynucleotides. Y1 cells were cultured in the presence of different concentrations of scrambled and antisense oligodeoxynucleotides (sequence shown in Materials and Methods) as indicated for 48 h. RNA (3 μg) extracted from the cells was subjected to an RNase protection assay as described (26) using a 700-bp riboprobe [probe A in Rouleau et al. (26)] encoding the DNA MeTase genomic sequence from −0.39 to +318. The major bands representing the two major initiation sites are indicated (92-, 90-bp, protected fragments) as well as the first exon, which gives a 99-bp, protected fragment. (B) Time course of inhibition of DNA MeTase mRNA by antisense oligodeoxynucleotides. Y1 cells were incubated in the presence of 20 μM of either antisense or scrambled oligodeoxynucleotides, and the medium was replaced with oligodeoxynucleotide-containing medium every 24 h. Cells were harvested at the indicated time points, and RNA and nuclear extracts were prepared as described. RNA was subjected to RNase protection assay as described in A. An autoradiogram similar to the one presented in A was scanned, and the amount of DNA MeTase mRNA at each point was normalized to the signal obtained for 18s ribosomal RNA. (C) Nuclear extracts prepared from oligodeoxynucleotide-treated Y1 cells described in B were assayed for DNA MeTase activity as described. The results represent an average of triplicate determination ± SD. (D) Y1 cells were treated with scrambled and antisense oligodeoxynucleotides as described in B and seeded onto soft agar for determination of anchorage-independent growth as described. The results represent an average of triplicate determinations ± SD.