Abstract
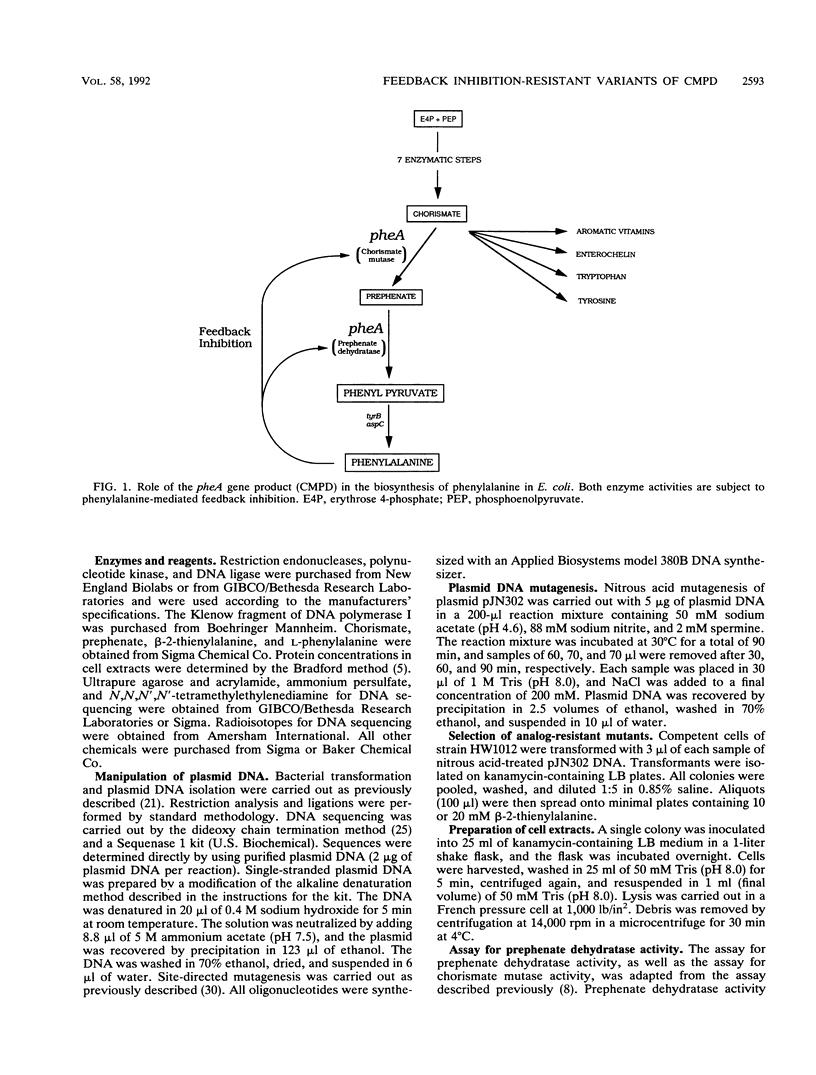
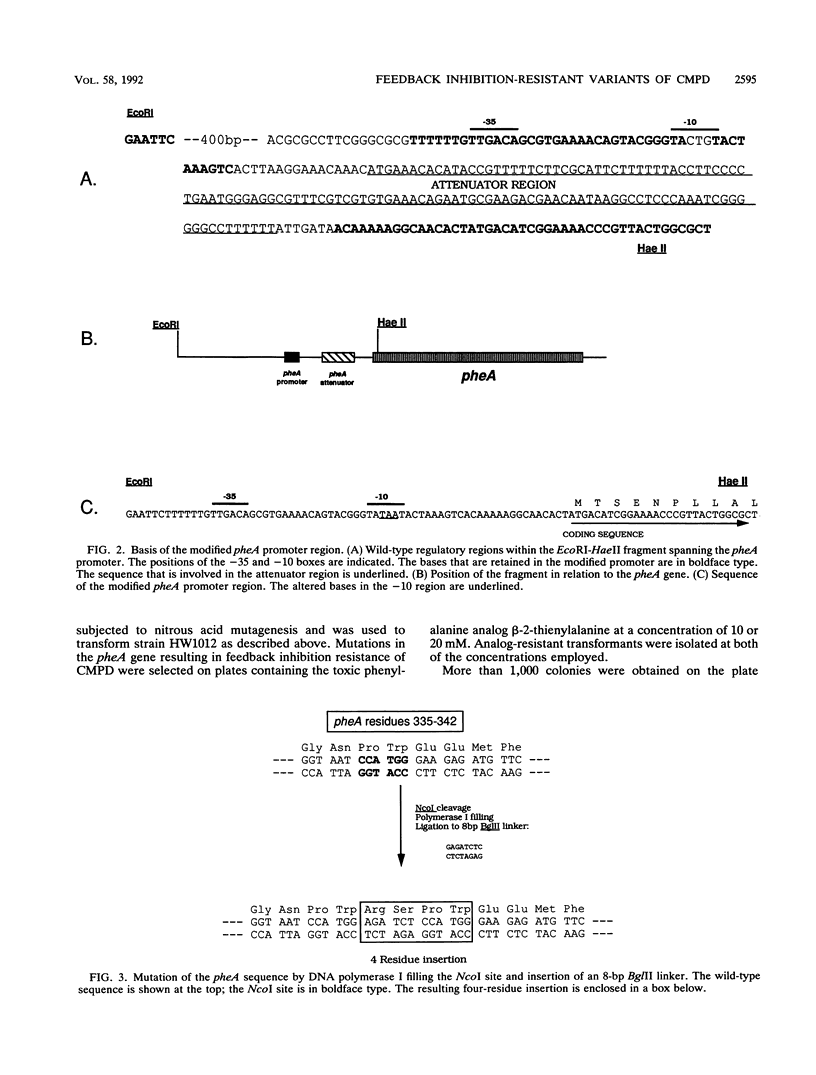
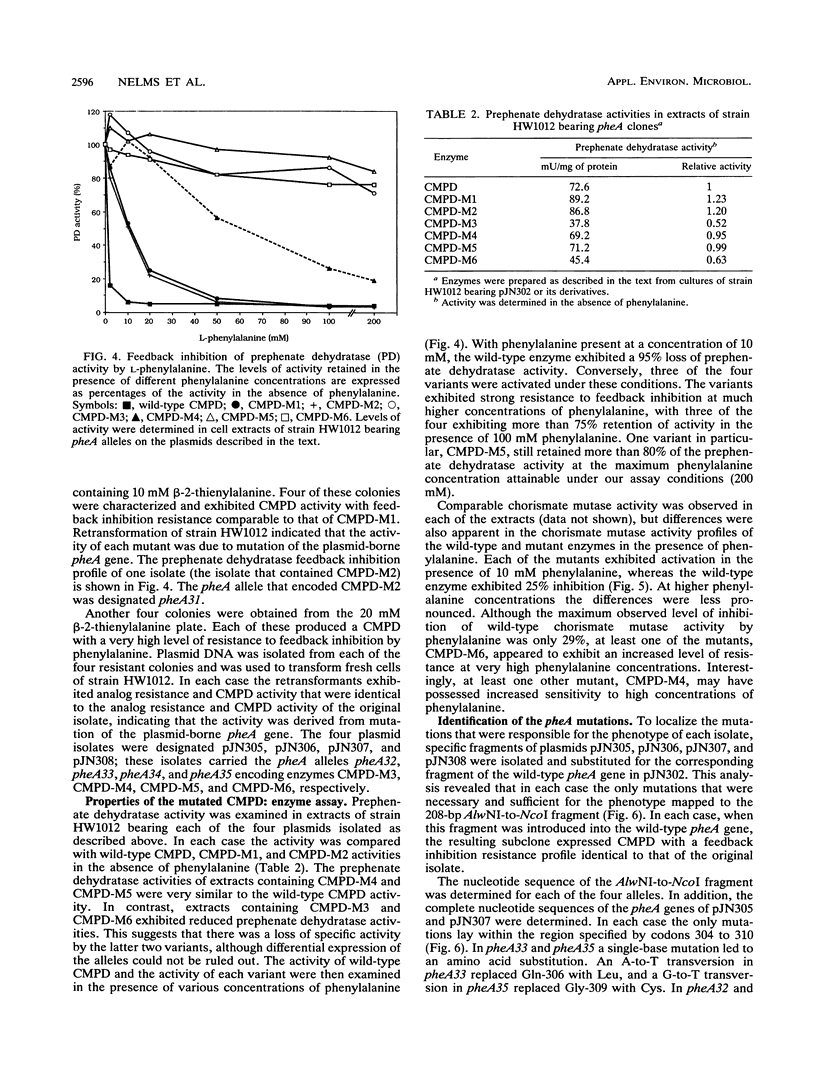
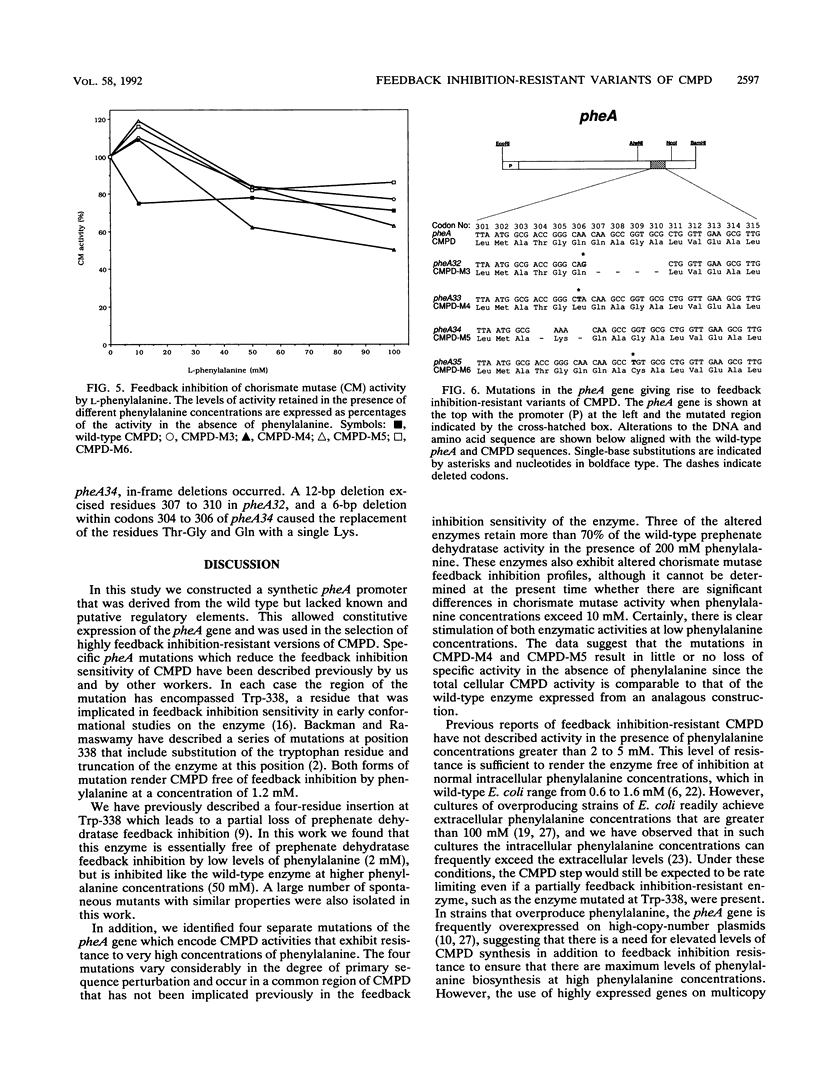
The bifunctional enzyme chorismate mutase/prephenate dehydratase (EC 5.4.99.5/4.2.1.51), which is encoded by the pheA gene of Escherichia coli K-12, is subject to strong feedback inhibition by L-phenylalanine. Inhibition of the prephenate dehydratase activity is almost complete at concentrations of L-phenylalanine greater than 1 mM. The pheA gene was cloned, and the promoter region was modified to enable constitutive expression of the gene on plasmid pJN302. As a preliminary to sequence analysis, a small DNA insertion at codon 338 of the pheA gene unexpectedly resulted in a partial loss of prephenate dehydratase feedback inhibition. Four other mutations in the pheA gene were identified following nitrous acid treatment of pJN302 and selection of E. coli transformants that were resistant to the toxic phenylalanine analog beta-2-thienylalanine. Each of the four mutations was located within codons 304 to 310 of the pheA gene and generated either a substitution or an in-frame deletion. The mutations led to activation of both enzymatic activities at low phenylalanine concentrations, and three of the resulting enzyme variants displayed almost complete resistance to feedback inhibition of prephenate dehydratase by phenylalanine concentrations up to 200 mM. In all four cases the mutations mapped in a region of the enzyme that has not been implicated previously in feedback inhibition sensitivity of the enzyme.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldwin G. S., McKenzie G. H., Davidson B. E. The self-association of chorismate mutase/prephenate dehydratase from Escherichia coli K12. Arch Biochem Biophys. 1981 Oct 1;211(1):76–85. doi: 10.1016/0003-9861(81)90431-8. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Brown K. D. Formation of aromatic amino acid pools in Escherichia coli K-12. J Bacteriol. 1970 Oct;104(1):177–188. doi: 10.1128/jb.104.1.177-188.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dopheide T. A., Crewther P., Davidson B. E. Chorismate mutase-prephenate dehydratase from Escherichia coli K-12. II. Kinetic properties. J Biol Chem. 1972 Jul 25;247(14):4447–4452. [PubMed] [Google Scholar]
- Gavini N., Davidson B. E. Regulation of pheA expression by the pheR product in Escherichia coli is mediated through attenuation of transcription. J Biol Chem. 1991 Apr 25;266(12):7750–7753. [PubMed] [Google Scholar]
- Gavini N., Davidson B. E. The pheR gene of Escherichia coli encodes tRNA(Phe), not a repressor protein. J Biol Chem. 1990 Dec 15;265(35):21527–21531. [PubMed] [Google Scholar]
- Gavini N., Davidson B. E. pheAo mutants of Escherichia coli have a defective pheA attenuator. J Biol Chem. 1990 Dec 15;265(35):21532–21535. [PubMed] [Google Scholar]
- Gelfand D. H., Steinberg R. A. Escherichia coli mutants deficient in the aspartate and aromatic amino acid aminotransferases. J Bacteriol. 1977 Apr;130(1):429–440. doi: 10.1128/jb.130.1.429-440.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gething M. J., Davidson B. E. Chorismate mutase/prephenate dehydratase from Escherichia coli K12. Effect of phenylalanine, NaCl and pH on the protein conformation. Eur J Biochem. 1978 May;86(1):159–164. doi: 10.1111/j.1432-1033.1978.tb12295.x. [DOI] [PubMed] [Google Scholar]
- Gowrishankar J., Pittard J. Regulation of phenylalanine biosynthesis in Escherichia coli K-12: control of transcription of the pheA operon. J Bacteriol. 1982 Jun;150(3):1130–1137. doi: 10.1128/jb.150.3.1130-1137.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson G. S., Davidson B. E. Nucleotide sequence and transcription of the phenylalanine and tyrosine operons of Escherichia coli K12. J Mol Biol. 1984 Dec 25;180(4):1023–1051. doi: 10.1016/0022-2836(84)90269-9. [DOI] [PubMed] [Google Scholar]
- Im S. W., Pittard J. Phenylalanine biosynthesis in Escherichia coli K-12: mutants derepressed for chorismate mutase P-prephenate dehydratase. J Bacteriol. 1971 Jun;106(3):784–790. doi: 10.1128/jb.106.3.784-790.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno J. R., Oxender D. L. Amino acid transport systems in Escherichia coli K-12. J Biol Chem. 1968 Nov 25;243(22):5914–5920. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoker N. G., Fairweather N. F., Spratt B. G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982 Jun;18(3):335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- Twigg A. J., Sherratt D. Trans-complementable copy-number mutants of plasmid ColE1. Nature. 1980 Jan 10;283(5743):216–218. doi: 10.1038/283216a0. [DOI] [PubMed] [Google Scholar]
- Zoller M. J., Smith M. Oligonucleotide-directed mutagenesis using M13-derived vectors: an efficient and general procedure for the production of point mutations in any fragment of DNA. Nucleic Acids Res. 1982 Oct 25;10(20):6487–6500. doi: 10.1093/nar/10.20.6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Brown K., Killingly D., Yanofsky C. Nucleotide sequence of the leader region of the phenylalanine operon of Escherichia coli. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4271–4275. doi: 10.1073/pnas.75.9.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]


