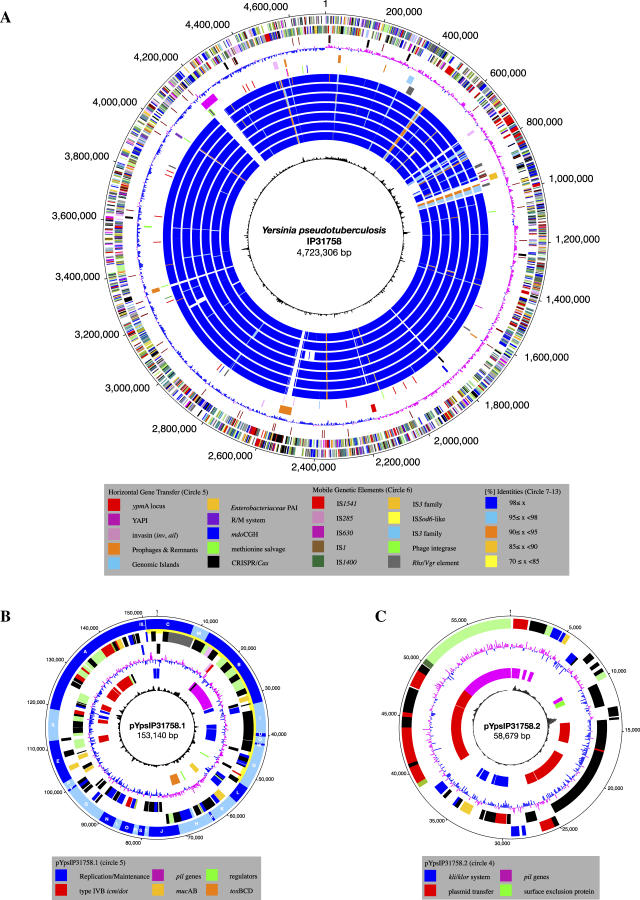
Figure 1. Circular Representation of the Y. pseudotuberculosis IP31758 Genome.
Chromosome (A), pYpsIP31758.1 (B), and pYpsIP31758.2 (C).
(A) Chromosome. Circles from outer to inner circle. (Circle 1–2) Predicted CDSs encoded on the plus (circle 1) and minus strands (circle 2), colored according to the respective TIGR role IDs (http://cmr.tigr.org/tigr-scripts/CMR/RoleIds.cgi). (Circle 3) Noncoding RNAs. Brown, tRNA genes; black, ribosomal rRNAs. (Circle 4) GC skew. (Circle 5) Genomic islands, including the YAPIIP31758 pathogenicity island, the Enterobacteriaceae PAI, strain-specific prophage insertions and phage remnants, plasmid-borne regions, and the superantigenic ypmA locus. Shared IP31758- and IP32953-specific R/M system with adjacent integrase. (Circle 6) IS elements, transposases, and phage integrases. (Circles 7–13) Comparative analysis of the genomic inventory. Intraspecies comparison of Y. pseudotuberculosis strains IP31758 and IP32953 (circle 7), interspecies comparison of Y. pseudotuberculosis IP31758 with Y. pestis strains CO92 (circle 8), KIM (circle 9), 91001 (circle 10), Nepal516 (circle 11), Antiqua (circle 12), and Pestoides F (circle 13). (Circle 14) Chi square.
(B) Plasmid pYpsIP31758.1. (Circle 1) HindIII restriction map. (Circles 2–3) Predicted CDSs encoded on the plus (circle 2) and minus (circle 3) strands, colored according to the respective TIGR role IDs (http://cmr.tigr.org/tigr-scripts/CMR/RoleIds.cgi). (Circle 4) GC skew. (Circle 5) Plasmid regions of interest. (Circle 6) Chi square.
(C) Plasmid pYpsIP31758.2. (Circles 1–2) Predicted CDSs encoded on the plus (circle 1) and minus (circle 2) strands, colored according to the respective TIGR role IDs. (Circle 3) GC skew. (Circle 4) Plasmid regions of interest. (Circle 5) Chi square.