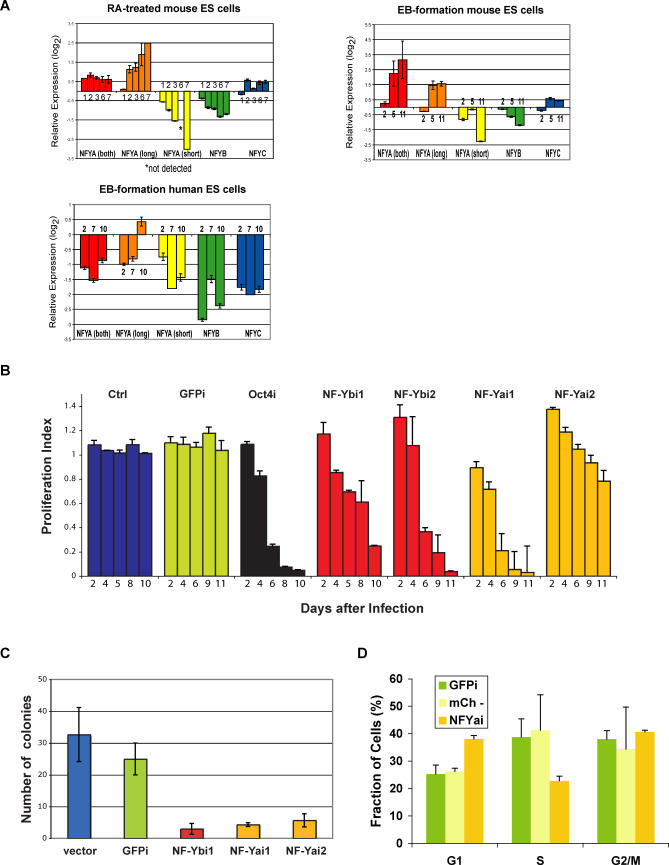
Figure 7. NF-Y Is Differentially Expressed during ES Cell Differentiation and Is Required for ES Cell Proliferation.
(A) Expression levels of NF-YA, NF-YA(long), NF-YA(short), NF-YB, and NF-YC during differentiation of ES cells. Real-time RT-PCR of RA-treated mouse ES cells, upper left panel; EB formation by mouse ES cells, upper right panel; EB formation by human ES cells, lower left panel. Fold-changes were calculated relative to undifferentiated ES cells using the REST software [94] and housekeeping genes as controls. A representative of at least three experiments (each performed in duplicate) is shown. Days of differentiation are indicated next to the bars.
(B) RNAi-based competition assay. Mouse ES cells were infected with a lentiviral vector that induces RNAi and labels the cells with a red fluorescent marker, mCherry. The percentage of cells undergoing RNAi (mCherry+) was measured in a competition assay with noninfected wild-type cells over time. The ratio [mCherry+ cells in RNAi against target gene/mCherry+ cells in RNAi against GFP] gives a proliferation index. In the case of cells undergoing RNAi against GFP (GFPi), the ratio was calculated using cells infected with empty lentiviral vector as control. This index is expected to remain at 1 over time if there are no effects of RNAi against the target gene on proliferation and to be less than 1 if there are defects in cell proliferation. We validated our approach with RNAi against Oct4 (black bars). Downregulation of NF-YA or NF-YB leads to defective proliferation of ES cells (orange and red bars, respectively), while the unrelated control sequence or downregulation of GFP has no effect (blue and green bars, respectively). Results from one of 3–5 independent experiments are shown. Bars represent averages of duplicates (NF-YB, control) or triplicates (NF-YA, GFP, Oct4) performed in a single experiment.
(C) Colony-formation assay. Control (vector and GFPi) cells and cells undergoing RNAi against NF-YA or B were sorted and plated at low density (300 cells per well of a 6-well plate) in triplicates. Colonies were counted after 10 d of culture.
(D) Cell-cycle analysis. Cells infected with a lentivirus leading to NF-YA knockdown were sorted for mCherry+ cells (NF-YAi), which are undergoing RNAi against NF-YA, or mCherry- cells (mCh-), which correspond to in-plate control noninfected cells with which the NF-YA knockdown ES cells are competing. An additional control used was ES GFPi cells. Cells were stained using propidium iodide and analyzed for DNA content using flow cytometry. Samples were analyzed in triplicates. Error bars depict standard deviation.