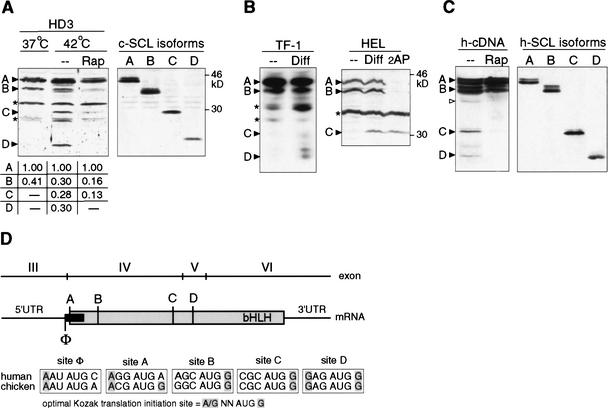
Figure 1.
Translational controlled differential expression of SCL protein isoforms during erythroid differentiation. (A) Erythroid differentiation in HD3 erythroblasts was induced by temperature shift (37°C–42°C for 12 h) to inactivate ts-v-erbB. (Left) Rapamycin was added 6 h after temperature shift where indicated. (Right) Expression of individual SCL isoforms from sequential translation initiation codons revealed by 5′ deletion of chicken SCL cDNA (termed A–D) in COS-1 cells. SCL-isoform expression was quantified by densitometry of X-ray films and normalized values are depicted below. For immunodetection, specific antiserum against C terminus of chicken SCL was used. (B) Human TF-1 or HEL cells were treated for 4–48 h with Epo or DMSO, respectively, to induce erythroid differentiation. 2-Amino purine (2AP) was added where indicated. Immunodetection of SCL in total cell extracts of TF-1 (data shown for 0 and 4 h postinduction) or HEL cells using a human SCL C terminus-specific antiserum is shown. In HEL cells, the SCL-D isoform was obscured by a nonspecific band and was therefore omitted. (C) Transiently transfected COS-1 cells with HA-tagged human SCL cDNA (left panel), and N-terminal-SCL deletion constructs (A–D, right panel). Cells were treated with rapamycin (Rap), or mock (− −) treated for 24 h where indicated. For immunodetection of SCL, an HA-immunotag-specific antiserum was used. SCL protein bands (▸), an alternative CUG (▹), and nonspecific/unidentified bands (*) are indicated. (D) Representation of the conserved vertebrate scl structure and translation initiation sites. Numbering of exons follows the nomenclature for the human scl gene. The SCL coding region is depicted in gray, the upstream open reading frame (uORF) is depicted in black. Potential translation initiation AUG-sites are designated A–D for SCL and Φ for the uORF. Shading in the initiation sites comparison indicates critical nucleotides at position −3 and +4 according to the optimal Kozak translation initiation consensus sequence (Kozak 1991).