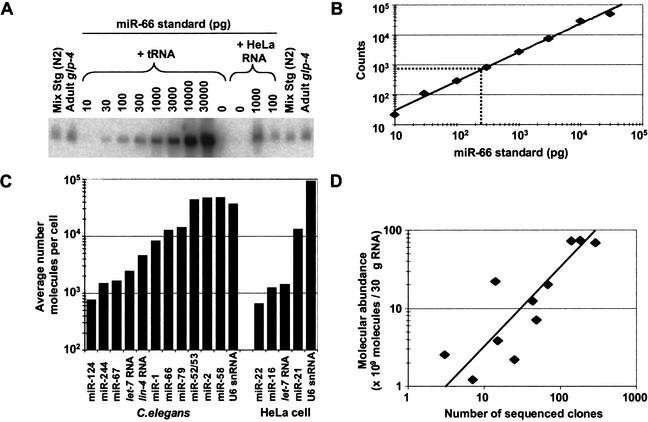
Figure 5.
Quantitative analysis of miRNA expression. (A) Northern blot used to quantify the abundance of miR-66. RNA prepared from the wild-type (N2) mixed-stage worms used in cloning and from glp-4(bn2) young adult worms were run in duplicate with a concentration course of synthetic miRNA standard. The signal from the standard did not change when total RNA from HeLa cells replaced E. coli tRNA as the RNA carrier, showing that the presence of other miRNAs did not influence membrane immobilization of the miRNA or hybridization of the probe. (B) Standard curve from quantitation of miR-66 concentration course. The best fit to the data is a line represented by the equation y = 3.3x0.96 (R2 = 0.99). Interpolation of the average signal in the glp-4 lanes indicates that the glp-4 samples contain 240 pg of miR-66 (broken lines). (C) Molecular abundance of miRNAs and U6 snRNA. Amounts of the indicated RNA species in the glp-4 samples were determined as shown in A and B. The average number of molecules per cell was then calculated considering the number of animals used to prepare the sample, and the yield of a radiolabeled miRNA spiked into the preparation at an early stage of RNA preparation. Analogous experiments were performed to determine the amounts of the indicated human miRNAs in HeLa RNA samples. (D) Correlation between miRNA molecular abundance and cloning frequency. The number of molecules in the mixed-stage RNA samples was determined as described for the glp-4 samples and then plotted as a function of the number of times the miRNAs was cloned from this mixed-stage population (Table 1). The line is best fit to the data and is represented by the equation y = 0.32x (R2 = 0.78).