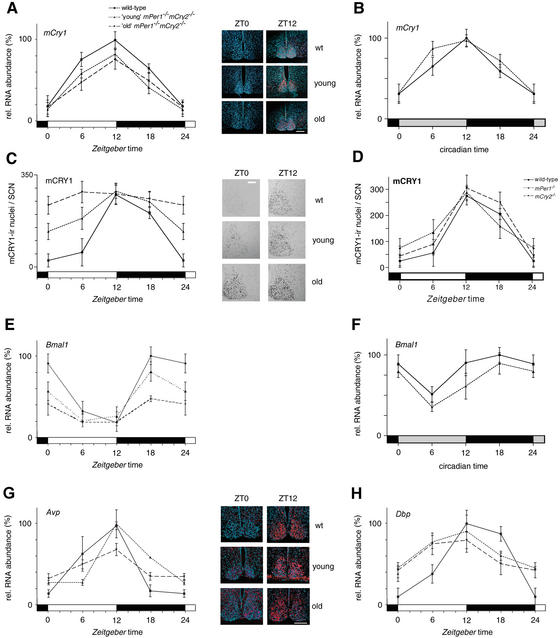
Figure 3.
mCry1 and Bmal1 mRNA, and mCRY1 protein expression profiles of wild-type (solid line), young mPer1-/- mCry2-/- (dotted line), and old mPer1-/- mCry2-/- (dashed line) mice. (A) Diurnal expression of mCry1 in the SCN in LD. The black and white bars on the X-axis indicate dark and light phases, respectively. (Right panels) Representative micrographs of SCN probed with the mCry1 antisense probe at time points of minimal(ZT0) and maximal(ZT12) expression. Bar, 200 μm. (B) Circadian expression of mCry1 in the SCN on the fourth day in DD. The gray and black bars on the X-axis indicate subjective day and night, respectively. (C) Diurnal variation of mCRY1 immunoreactivity in the SCN in LD. Quantification was performed by counting immunoreactive nuclei in the area of the SCN. In old double mutants, oscillation of mCRY1 immunoreactivity is significantly dampened (p < 0.05), with constantly high numbers of immunoreactive nuclei throughout the LD cycle. (Right panels) Representative micrographs of immunostained SCN at time points of minimal(ZT0) and maximal(ZT12) immunoreactivity. Bar, 100 μm. (D) Diurnal expression of mCRY1 protein in the SCN of wild-type (solid line), mPer1-/- (dotted line), and mCry2-/- (hatched line) mice. (E) Diurnal variation of Bmal1 mRNA expression in the SCN in LD. In old double mutants, Bmal1 cycling is significantly dampened (p < 0.05). (F) Circadian expression of Bmal1 mRNA in the SCN on the fourth day in DD. (G) Diurnal time course of Avp mRNA expression in the SCN in LD. (H) Diurnal time course of Dbp mRNA expression in the SCN in LD. Tissue was visualized by Hoechst dye nuclear staining (blue); silver grains are artificially colored (red) for clarification. All data presented are mean ± S.D. for three different experiments.