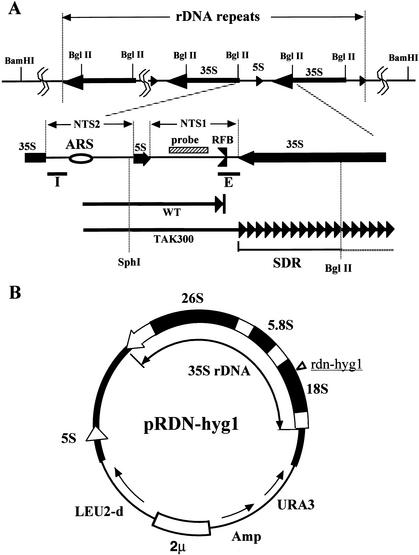
Figure 1.
(A) Structure of rDNA repeats in Saccharomyces cerevisiae. The locations of the 35S and 5S rRNA genes (the direction of transcription is indicated by arrows) are shown in the upper part. The recognition sites of BglII are also shown. The two nontranscribed spacer regions (NTS1 and NTS2), the ARS (replication origin), and its surrounding regions are expanded below. The RFB (replication fork barrier site) is indicated. The striped box is the region used for the rDNA-specific probe. The solid bars I and E are the components of HOT1. Two arrows in the bottom part show progression of one of the replication forks started from the ARS, bidirectionally. The upper fork (WT, wild type) is arrested at the RFB, and the lower one (TAK300; fob1, low rDNA copy number) is slowed down in the 35S rDNA. SDR is the region in which the replication fork is slowed down (Fig. 2C, panel c). The recognition sites of SphI and BglII, which were used for the 2D gel analysis (Fig. 2C), are also shown. (B) Structure of the helper plasmid, pRDN-hyg1 (Chernoff et al. 1994). The filled box shows an rDNA unit. The hygromycin B-resistance mutation site is indicated in the 18S rDNA. This plasmid was used to isolate strains with low copy numbers of rDNA repeats.