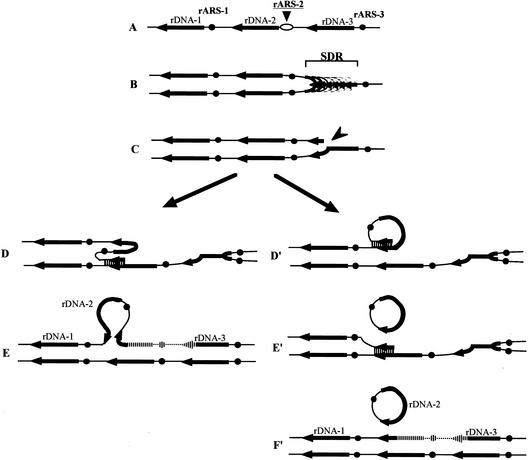
Figure 6.
The transcription-dependent recombination model for rDNA repeat expansion, contraction, and production of ERCs in TAK300 (low rDNA copy number, fob1). The positions of the ARS and the 35S rDNA are shown as filled dots and arrows, respectively. Individual lines represent chromatids with double-stranded DNA. In this model, DNA replication starts from one of the ARSs (ARS-2) bidirectionally (A). As all of the 35S rDNA units are transcribed fiercely in this strain, the rightward replication fork is slowed down in the 35S rDNA (SDR; B), and we propose that it stimulates a DNA double-strand break (indicated by an arrowhead in C). As the strain is fob1, the replication fork barrier (RFB) site at the 3′-end of the 35S rDNA is not functioning. (D) A strand invasion at a homologous duplex (a downstream sister chromatid near ARS-1 in this example) takes place, and a new replication fork is formed. (E) The new replication fork meets with the leftward replication fork, resulting in formation of two sister chromatids, one of which gains an extra copy of rDNA, indicated as a striped line. If the strand invasion is at a site in an upstream repeat (e.g., right side of ARS-3), a loss, rather than a gain, of an rDNA repeat is expected. If the strand invasion is at a site in the same chromatid (D′), an ERC is produced (E′). Then another invasion takes place (E′, lower part) and two sister chromatids are formed (F′). The region that is replicated twice is indicated as a striped line in F′. A similar model that does not involve transcription-related ERC production is presented by Rothstein and Gangloff (1999).