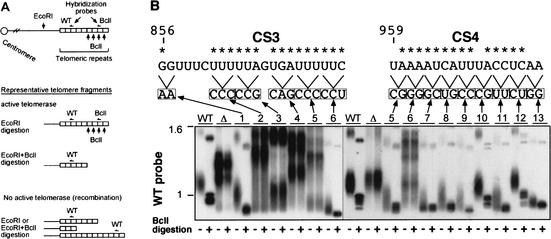
Figure 2.
Sequences CS3 and CS4 in K. lactis telomerase RNA are important for telomerase action and telomere maintenance in vivo. (A) Schematic showing a telomeric restriction fragment, and digests and probes used to analyze telomeres in the Bcl-marked system for telomerase RNA mutants. Also shown on the map are the positions of wild-type (WT) and Bcl-specific probes. (B) Southern blotting analysis of telomeric restriction fragments in K. lactis expressing different CS3- and CS4-region-scanning substitution mutants. Genomic DNA was prepared from K. lactis strains (deleted for the chromosomal TER gene and carrying the different TER alleles on a plasmid; see bottom part of figure) at their sixth passage. The control strains carry the phenotypically wild-type, BclI-marked TER gene (WT), or no insert (Δ) on a plasmid (Tzfati et al. 2000). DNA was digested with EcoRI restriction endonuclease (- lanes) or double-digested with EcoRI and BclI (+ lanes), separated on a 1% agarose gel, and vacuum-blotted. Hybridization was carried out as described previously (Tzfati et al. 2000), first with a BclI-specific oligonucleotide probe (data not shown), and then with a wild-type telomeric sequence probe. For simplicity, only the portion of the gel with a group of seven similarly sized telomeric EcoRI restriction fragments (McEachern et al. 2002) in WT K. lactis is shown. The positions of the scanning substitution mutations tested in the CS3 and CS4 regions are indicated above the autoradiograph. Asterisks indicate conserved residues (see Fig. 1A).