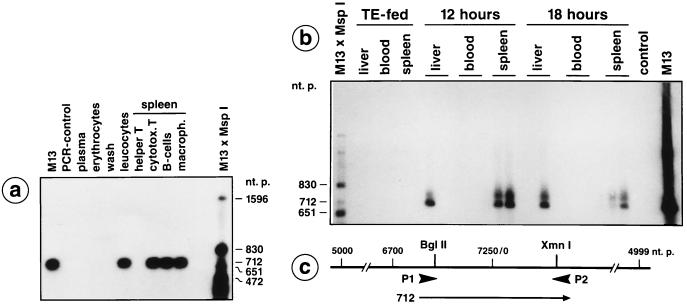
Figure 2.
Detection of M13mp18 DNA fragments in peripheral blood leukocytes, in liver, and in different spleen cells by PCR. (a) A male C57BL/6 mouse was pipette-fed 50 μg of M13mp18 DNA. At 4 h after feeding, blood was drawn by heart puncture and the plasma, erythrocyte, and leukocyte fractions were prepared by Ficoll gradient centrifugation. The leukocytes were washed twice in Tris-saline or PBS and incubated for 2 min in 0.15 M NaCl, 10 mM Tris·HCl (pH 7.5), 2 mM MgCl2, 1% Nonidet P-40. The thus liberated nuclei were sedimented, resuspended in 10 mM Tris·HCl (pH 7.5), 1 mM EDTA, 1% SDS, 1.3 mg/ml proteinase K, and incubated for 2 h at 37°C. Nucleic acids were then prepared by phenol/chloroform extraction. Similarly, the plasma, erythrocyte, and wash fractions were extracted for possibly present DNA. By using the synthetic oligodeoxyribonucleotide primers P1 and P2 designated on the M13mp18 DNA map in c, the indicated M13mp18 DNA segments were amplified as described earlier (1). (a) At 18 h after feeding a different animal, the different spleen cells were fractionated by the magnetic bead method as described. DNA from the designated cell fractions was prepared and PCR analyzed. (b) Detection of M13mp18 DNA in spleen and liver cells by PCR at 12 or 18 h after feeding. DNA samples from TE-fed animals or from blood of animals 12 or 18 h after feeding M13mp18 DNA did not contain M13mp18 DNA. The PCR products were blotted to positively charged membranes, and M13mp18 DNA was identified by hybridization to 32P-labeled M13mp18 DNA and by autoradiography.