Abstract
Members of the POU and SOX transcription factor families exemplify the partnerships established between various transcriptional regulators during early embryonic development. Although functional cooperativity between key regulator proteins is pivotal for milestone decisions in mammalian development, little is known about the underlying molecular mechanisms. In this study, we focus on two transcription factors, Oct4 and Sox2, as their combination on DNA is considered to direct the establishment of the first three lineages in the mammalian embryo. Using experimental high-resolution structure determination, followed by model building and experimental validation, we found that Oct4 and Sox2 were able to dimerize onto DNA in distinct conformational arrangements. We demonstrate that the DNA enhancer region of their target genes is responsible for the correct spatial alignment of glue-like interaction domains on their surface. Interestingly, these surfaces frequently have redundant functions and are instrumental in recruiting various interacting protein partners.
Keywords: Oct4, Sox2, POU domain, HMG domain, FGF4 and UTF1 enhancers, crystal structure
A surprisingly low number of transcription factors is responsible for regulating the development of an entire organism. These transcription factors form multiprotein complexes on DNA, thereby orchestrating the correct temporal–spatial expression of developmental genes. The process leads to the establishment of functional partnerships, with the combination rather than the individual activity of each factor eliciting specific transcriptional outcomes. Members of the transcription factor families POU and SOX exemplify this functional cooperativity during early embryonic development. POU (Herr and Cleary 1995) and SOX (Wegner 1999) proteins selectively interact with each other via the conserved domains POU and HMG, respectively, which also bind to DNA. Their functional partnership has been characterized on regulatory elements in various species, including human, mouse, and the fruit fly (Dailey and Basilico 2001). POU and SOX proteins are differentially expressed during development, and their combinations may lead to the differential expression of genes critical for cell-fate determination (Dailey and Basilico 2001). The genes encoding the transcription factors Oct4 and Sox2 are tightly regulated during development and in embryonic cell lines (see Discussion). Their combination is critical, as it functions to specify the first three lineages in the mammalian embryo (Nichols et al. 1998; Niwa et al. 2000; Avilion et al. 2003). Whereas Oct4 and Sox2 are considered to define a combinatorial code in vivo (Avilion et al. 2003), binding of POU factors by Sox2 in vitro is rather indiscriminate. For example, the POU domains of several family members, including the prototype member Oct1, bind cooperatively with the HMG domain of Sox2 onto the fibroblast growth factor 4 (FGF4) enhancer (Ambrosetti et al. 1997). This DNA regulatory element contains an octamer motif for POU binding and an adjacent motif for HMG binding. However, its activation in vivo is dependent on Sox2/Oct4 binding and is mediated by Oct4-specific regions external to the POU domain (Ambrosetti et al. 2000). Another POU/SOX-dependentelementis responsible for regulating expression of the Undifferentiated Transcription Factor 1 gene (UTF1), which contains a modified octamer motif (AT GCTAGT) and a SOX-binding site (Nishimoto et al. 1999).
In this study, we investigate the interaction of Oct1 and Oct4 with Sox2 on two different DNA enhancers to test whether a previously discovered regulation mechanism of DNA-mediated swapping of the arrangement of homodimers (Tomilin et al. 2000; Reményi et al. 2001b) may also be applicable for unrelated transcription factor assemblies. We first solved the crystal structure of the ternary Oct1/Sox2/FGF4 enhancer elementcomplex and then used homology modeling tools to construct an Oct4/Sox2/FGF4 as well as an Oct4/Sox2/UTF1 structural model. These models revealed that the FGF4 and the UTF1 enhancers mediate the assembly of distinct POU/HMG complexes, leading to different quaternary arrangements by swapping protein–protein interaction surfaces of Sox2. Moreover, we demonstrate that Sox2 uses one of its two protein interacting surfaces to assemble a ternary complex with another unrelated transcription factor on a late-embryonic-stage-specific enhancer (Pax6/Sox2 on the DC5 element). Our findings outline a simple mechanism for promiscuous yet highly specific assembly of transcription factors, in which the sequence of DNA enhancers governs a combinatorial use of redundant protein–protein interaction surfaces.
Results
Oct4 and Sox2interact differentially on the FGF4 and UTF1 enhancers
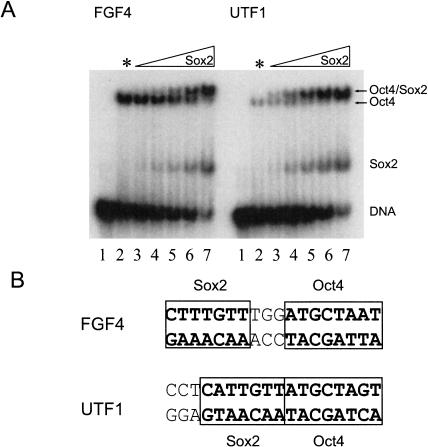
A comparative titration of Sox2 with Oct4 on the FGF4 and UTF1 enhancers in an electrophoretic DNA mobility shift assay (EMSA) revealed that in vitro, Oct4 and Sox2 interact with each other differently on these two enhancers. A lower amountof Sox2 HMG domain was required for heterodimerization with Oct4 on UTF1 than on FGF4 (Fig. 1A), the Oct4 POU domain being sufficient to exert this differential cooperativity (data not shown).
Figure 1.
Oct4 and Sox2 differentially interact on FGF4 and UTF1. (A) EMSA assay of Oct4 and Sox2 with radiolabeled DNA oligonucleotides of FGF4 and UTF1. (Lane 1) No protein, DNA alone. (Lane 2) Oct4 alone + DNA (* indicates that no Sox2 protein was added to this lane). (Lanes 3–7) Increasing amounts of Sox2-HMG protein mixed with equal amounts of Oct4 + DNA. Although Oct4 binds alone to UTF1 significantly more weakly than to FGF4 (cf. the two lane 2s), heterodimerization on UTF1 is more pronounced with even lower amounts of Sox2 (e.g., cf. the two lane 4s or 5s). (Remark: Sox2 binds FGF4 and UTF1 with similar affinity; see Supplemental Material.) The significantly lower degree of Oct4/DNA interaction in the absence of Sox2 on UTF1 compared with that on FGF4 is very likely caused by the nonoptimal octamer motif sequence for POU binding within this DNA element (Nishimoto et al. 1999). (B) Differential spacing of binding sites for Oct4 and Sox2 in FGF4 and UTF1.
The FGF4 enhancer contains 3 bp between the POU- and HMG-binding sites, but no such spacer is present between the respective sites within the UTF1 enhancer (Fig. 1B). We were therefore interested in whether the observed different biochemical properties of the POU/HMG complexes formed on the two enhancers could be attributed to this different spacing of the binding sites, similar to the earlier example of POU factor dimerization (Tomilin et al. 2000; Reményi et al. 2001b). In our previous studies, we have shown that the transcriptional activity of POU factor dimers is regulated by alterations in their quaternary arrangement, which are induced by binding to specific regulatory elements of target genes (Tomilin et al. 2000; Reményi et al. 2001b).
Crystal structure determination of the Oct1/Sox2/FGF4 ternary complex
First, the crystal structure of the POU/HMG ternary complex on the FGF4 enhancer was determined using the POU domain of Oct1 and the HMG domain of Sox2. The crystal structure was solved by the MAD method (Table 1). POU factor homodimerization on two different elements, PORE and MORE, has been characterized structurally with the POU domain of Oct1, and it represents an example analogous to POU/SOX heterodimerization: differential transcriptional activity could be achieved by the same set of transcription factors, provided that they interact on distinct elements. We chose to solve a POU/SOX/DNA structure with the POU domain of Oct1 rather than with Oct4 because Oct1 is regarded as the prototype member of the POU transcription factor family. Moreover, the POU domain of Oct1 is ∼60% identical to that of Oct4 and has previously been shown to form a cooperative ternary complex with the HMG domain of Sox2 (Ambrosetti et al. 1997). Furthermore, consistently using Oct1 for structural studies allows us to directly compare POU factor homodimerization and POU/SOX heterodimerization on different DNA elements. The FGF4 enhancer was the first DNA element that was described to contain a composite DNA elementbinding Sox2 and Oct4 (Yuan et al. 1995). Further biochemical work demonstrated the cooperative nature of this interaction with Oct4 as well as with Oct1 (Ambrosetti et al. 1997). Therefore, we chose the Oct1/Sox2/FGF4 ternary complex as the crystallization target.
Table 1.
Crystallographic data collection, structure solution, and refinement
| Crystal [space group] | λ (Å) | d (mi) (Å) | No. data | Completeness (%) | Multiplicity | I/σ (last shell) | Rsymb (%) | Phasing powerc iso/ano | Rcullisd iso/ano |
|---|---|---|---|---|---|---|---|---|---|
| X-ray data collection | |||||||||
| P/H:F23ta [P3121] | 0.8460 | 2.6 | 16,619 | 98.3 | 5.2 | 21.8 (2.5) | 4.7 (35.0) | — | — |
| P/H:F22ta (br)-peak [P6422] | 0.9198 | 3.0 | 24,450a | 99.6 | 5.5 | 17.0 (2.4) | 4.3 (53.2) | —/1.63 | —/0.79 |
| P/H:F22ta (br)-infl. | 0.9202 | 3.0 | 24,366a | 99.5 | 4.3 | 19.2 (2.0) | 3.9 (55.1) | 3.02/1.08 | 0.47/0.92 |
| P/H:F22ta (br)-high | 0.9068 | 3.0 | 22,662a | 92.2 | 2.9 | 15.1 (1.7) | 4.5 (56.8) | 0.98/2.05 | 0.61/0.82 |
| Crystal | Resolution (Å) | Protein atoms | DNA atoms | Solvent atoms | RMSd bond length (Å) | RMSd bond angles (°) | Rcryste (%) | Rfreee (%) | <B> (Å2) protein/DNA |
| Structure refinement | |||||||||
| P/H:F23ta | 30.0-2.6 | 1765 | 978 | 90 | 0.010 | 1.8 | 23.2 | 28.5 | 61/54 |
Friedel mates were not merged during scaling.
Rsym = ΣhklΣi|Ii (hkl) - <I(hkl)>|/ΣhklΣI Ii(hkl).
Phasing power is defined as the ratio of the RMS value of the heavy atom structure factor amplitudes and the RMS value of the lack-of closure error.
Rcullis is the mean lack-of-closure error divided by the isomorphous/anomalous difference.
Rcryst and Rfree = |ΣFobs - Fcalc|/ΣFobs; Rfree is calculated with 5% of the data that were not used for refinement.
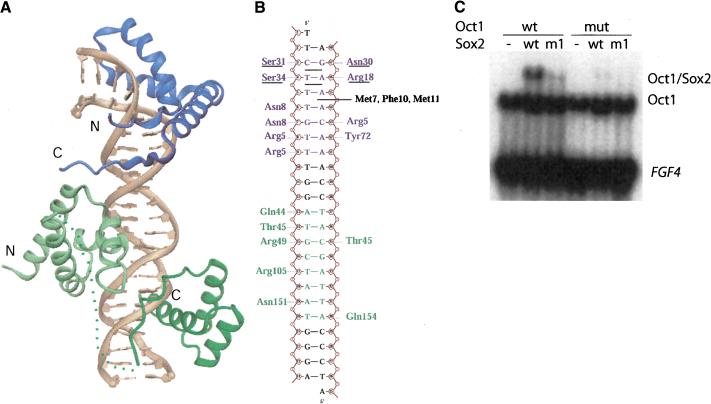
The crystal structure of the Oct1/Sox2/FGF4 ternary complex reveals a novel heterotrimeric domain arrangement, in which the centrally positioned POU-specific domain (POUS) interacts with the HMG domain of Sox2 and the POU homeodomain (POUH) of Oct1 (Fig. 2A). The Sox2 HMG domain adopts an L-shaped structure, and its N-terminal 70-residue segment folds like other structurally characterized HMG domains (Murphy and Churchill 2000).
Figure 2.
Crystal structure of the Oct1/Sox2/FGF4 ternary complex. (A) The centrally positioned POU-specific domain (POUS) interacts with the HMG domain of Sox2. The HMG domain of Sox2 is blue and the POU domain of Oct is green (POUs: light green; POUH; dark geeen). Some part of the POU linker region connecting POUs and POUH is invisible in the crystal structure (shown as a dotted line). (B) Protein—DNA interactions between the HMG, POU, and the FGF4 enhancer. The figure shows the sequence of the oligonucleotide used for crystallization. The HMG and the POU domain binding sites are blue and green, respectively. For simplicitty, only DNA-sequence-specific hydrogen bonds are shown. The hydrophobic side chains of M7, F10, and M11 are inserted between base pairs T5 · A45 and T6 · A44, causing an ∼45° bend of the DNA axis. These and other amino acid residues that play a role in bending the DNA at three different base stack levels are either underlined or written in black. (C) EMSA results of Octl/Sox2/DNA ternary complex formation on the FGF4 enhancer using mutant versions of the HMG domain of Sox2 (m1) and POU domain of Oct1 (mut). The mutants were designed to interfere with the POU-HMG interface formation based on the Oct1/Sox2/FGF4 crystal structure. The R75E mutation in Sox2 (m1) and the I21Y, D29R mutation in Oct1 (mut) were introduced to reverse the charges or increase the bulkiness of important amino acids at the POU/HMG interface. (wt) Wild-type protein.
Sox2-HMG/DNA interaction
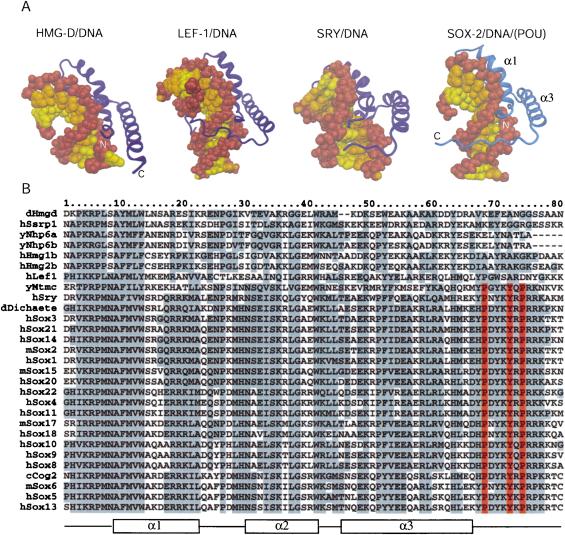
The first 70 residues of Sox2 superimpose with other HMG domains from HMG-D, Lef-1, and Sry with an RMSD of 4.0, 1.7, and 3.4 Å, respectively (Love et al. 1995; Werner et al. 1995; Murphy et al. 1999). Sox2 binds in the minor groove of the DNA and forms an HMG/DNA interaction surface that is comparable in size to that of the POU/DNA (1350 Å2 and 1400 Å2, respectively; Fig. 2B). The HMG domain severely bends the DNA toward the major groove with an approximate bend angle of 90°. Side chains from residues of helix 1 and helix 2 of the Sox2-HMG domain are inserted between 3-bp stacks of the recognition sequence (C^T^T^TGTT; Fig. 2B), leading to unwinding of the DNA helix at the Sox2-binding site. Consequently, the minor groove becomes shallow and expanded with an average groove width of 12 Å and negligible depth between C3 · G47 and T6 · A44 (calculated by CURVES; Lavery and Sklenar 1988; see Supplemental Material). Bending and opening of the minor groove is complemented by the compression of the major groove between T6 · A44 and G7 · C43. The minor groove between T9 · A41 and G11 · C39 is relatively narrow and deep with an average width and depth of ∼5–6 Å. Interestingly, the C-terminal tail of the Sox2-HMG domain snugly fits into this narrowed minor groove in between the Sox2- and the POU-binding sites.
Sox2 interacts with DNA in a sequence-specific manner and this specificity is mediated by numerous base-pair-specific hydrogen bonds (Fig. 2B). The Sox2/DNA interaction pattern is most closely related to that of the Sry/DNA complex (Werner et al. 1995). For instance, Asn 8, Ser 31, Ser 34, and Tyr 72 (Sox2 numbering) from Sox2 and Sry are involved in the same sequence-specific protein–DNA interactions. Some of the observed differences between the two proteins (e.g., Arg 5 and Asn 30) could be responsible for their slightly different DNA site preferences (Mertin et al. 1999). Whereas these two proteins bind DNA sequence-specifically, HMG-D, for example, interacts with DNA in a non-sequence-specific manner. DNA-interface residues that are conserved among SOX family members and Sry are diverse in other HMG domains (see Fig. 3B).
Figure 3.
Comparison of differentHMG domains. (A) Structures of different HMG domains from HMG-D (Murphy et al. 1999), Lef-1 (Love et al. 1995), Sry (Werner et al. 1995), and Sox2 (taken from the Oct1/Sox2/FGF4 crystal structure) in complex with DNA. Sox2 generates a similar bend angle (90°) in DNA as reported for other HMG/DNA complex structures (HMG-D, 111°; Lef-1, 117°; Sry, 75°). Helix 1 and helix 2 form extensive contacts with the DNA in the widened minor groove and are involved in bending the DNA in all structures. However, the proteins show significant differences in how their C-terminal regions interact with the DNA molecule. In HMG-D, which binds DNA without sequence specificity, only helices 1 and 2 interact with the DNA. The C terminus of the Lef-1 HMG domain lies in the compressed major groove and stabilizes the bent DNA conformation. In the Sry/DNA structure, the C-terminal part is mainly disordered and is not positioned in the minor groove. The Sox2-HMG C-terminal region fits tightly into the compressed minor groove when in the presence of the POUS domain. The DNA sugar-phosphate backbone is brown, and the bases belonging to different chains of the DNA molecule are depicted by differentcolors (yellow and orange). Helix 2 (α2) is behind the DNA molecule and, therefore, cannot be seen from this orientation. (B) Multiple sequence alignment of HMG domains of several proteins from differentorganisms. (y) Yeast; (c) Caenorhabditis elegans; (d) Drosophila melanogaster; (m) mouse; (h) human. The order of the proteins on the list reflects their sequence similarity. HMG domains from the first six proteins (dHmgd–hHmg2b) are known to bind DNA in a non-sequence-specific manner, whereas the others (hLef1–hSox13) bind DNA according to a specific sequence. Secondary structure elements of Sox2 from the Oct1/Sox2/DNA crystal structure are shown beneath the alignment. Protein residues that are highly conserved are boxed in gray. It is notable that the protein residues that play an important role in the ordering of the C-terminal region of the HMG from Sox2 (V3, R5, P6, H63, H67, P68, Y70, Y72, R75, and R76) are conserved in almostall members of the SOX family. However, these residues are divergent in HMG domains that bind DNA in a non-sequence-specific manner (Murphy and Churchill 2000). Residues P68, Y72, and P74, which play the most prominent role in positioning the C terminus in the compressed minor groove (cf. Fig. 4A), are red.
The POU/HMG interface
In contrast to previously determined HMG structures, the C terminus of the HMG domain (residues 68–79) in the Oct1/Sox2/FGF4 complex is ordered and bound in an extended β-strand-like conformation in the compressed minor groove located between the HMG and the POU-domain DNA-binding sites (Fig. 3A). This C-terminal segment interacts with the DNA and also forms a protein–protein interface by contacting a loop of the Oct1 POUS domain between helices 1 and 2. There is only one sequence-specific interaction between the two domains, a saltbridge between R75 (HMG) and D29 (POUS; see Fig. 2C), which could provide a rationale for the observed indiscriminate nature of POU/HMG/DNA complex formation (Ambrosetti et al. 1997). Thus, the structure of the Oct1/Sox2/FGF4 complex suggests that the ordering of the C-terminal part of the Sox2-HMG domain is induced by the presence of a POU/HMG interface. A multiple sequence alignment (see Fig. 3B) reveals that this HMG C terminus (residues 68–79) is virtually identical among all SOX members of the HMG family, but unrelated in sequence to members of other HMG subfamilies. This suggests that heterodimer interface formation via the HMG domain C terminus is a property limited to the SOX subgroup of HMG proteins. Moreover, the complementary POUS surface patch is also highly conserved among POU factors (Herr and Cleary 1995). The C terminus of the Sox2-HMG domain, which is presumably unstructured in the absence of an interacting protein partner, is likely to be a major contributor to ternary complex formation, because the interaction of this portion of the protein with the compressed minor groove increases the HMG protein/DNA surface by about one-third of the total (420 Å2/1350 Å2). Sox2 is able to bind to DNA on its own, but with a significantly lower affinity compared with binding to DNA as part of a ternary complex with POU or PAX proteins. This observation is in agreement with the supposition that SOX proteins are converted into high-affinity ligand binders in the presence of other DNA-binding protein partners (Kamachi et al. 2000).
Homology modeling of Oct4/Sox2/DNA ternary complexes
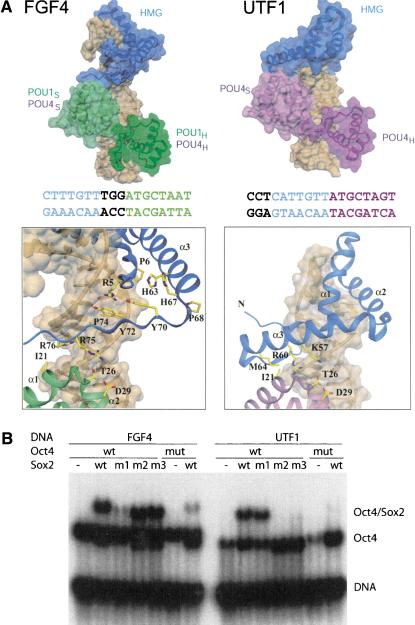
The Oct1/Sox2/FGF4 crystal structure allowed us to generate both a reliable Oct4/Sox2/FGF4 model based on homology (Vriend 1990), and to build an Oct4/Sox2/UTF1 ternary complex model (Fig. 4A). The latter was created by using the experimental structure of the Oct1/Sox2/FGF4 complex and keeping the Sox2-HMG domain and the Oct1/Oct4-POU domains (POU1 and POU4) at their consensus-like DNA sequences. Because Sox2 and POU factors bind DNA in a highly sequence-specific manner, their positioning on composite DNA sites can be reliably inferred (see Fig. 1B). The new model is based on a rotational movement of the HMG domain toward POUS, and suggests a second POU-HMG interface. In both arrangements, the same surface patch of the POUS subdomain is used. In contrast, the HMG domain interacts via a segment of helix 3 (K57–M64) on UTF1 instead of its C terminus as on FGF4, thus changing from an extended β-strand-like structure to an α-helical interface.
Figure 4.
Comparison of POU/HMG complexes formed on FGF4 and UTF1. (A, top) Model of POU/HMG/FGF4 (POU domain of Oct1 or Oct4; left), compared with the model of Oct4-POU/HMG/UTF1 (right). The figure illustrates that different spacing between the binding sites for the POU and HMG domains within FGF4 and UTF1 enhancers causes formation of different heterodimeric interfaces. (Bottom) Close-up views of the HMG/POUS interfaces on FGF4 (left) and UTF1 (right). The POU/HMG/UTF1 model suggests involvement of helix 3 instead of the C terminus of the HMG domain to form the HMG-POUS interface, whereas the same surface patch of the POUS domain appears to be involved in both interfaces. The DNA molecules are depicted with a transparent surface and are brown. Coloring of the FGF4 and UTF1 DNA sequences is according to their POU- and Sox2-HMG-binding sites. Notice the difference in spacing of these two sites in the two enhancers. (B) To validate the homology models in A, mutations in Sox2-HMG were designed to selectively interfere with ternary complex formation on the FGF4 (m1) or on the UTF1 enhancers (m2 and m3); (m1) R75E; (m2) K57E,R60E; (m3) R60E,M64E; (mut) mutant version of Oct4 POU (I21Y,D29E). In agreement with both models, m1 specifically impaired heterodimerization on FGF4, whereas m2 and m3 specifically abrogated heterodimerization on UTF1, but Oct4 mut affected both.
Sox2has two surface patches for interaction with Oct4
To test whether, indeed, these two distinct protein interfaces exist on the Sox2-HMG protein, we introduced three mutations in its C-terminal region: R75E (FGF4-specific; m1), K57E, R60E (UTF1-specific; m2) and R60E, M64E (UTF1-specific; m3). We found that none of the mutations had a significant impact on Sox2/DNA interaction, as the individual proteins bound to DNA similar to the respective wild-type protein (see Supplemental Material). The proteins were then analyzed for heterodimer formation with Oct4 in EMSA (Fig. 5B). The three mutants showed differential ternary complex formation with the FGF4 and UTF1 elements. Although both FGF4 and UTF1 mediated assembly of a ternary complex with the POU domain of Oct4 and wild-type Sox2-HMG, heterodimerization with either m2 or m3 on UTF1 and with m1 on FGF4 was selectively compromised. I21 and D29 of the POU domain play a prominent role in protein–protein interaction with HMG in the POU/HMG/FGF4 as well as in the POU/HMG/UTF1 complex. In agreement with this observation, the I21Y, D29R double mutation in Oct4 disrupted the ternary complex formation on both elements. In summary, these results validated the proposed models of Oct4/Sox2/DNA complexes with the FGF4 and UTF1 regulatory elements.
Figure 5.
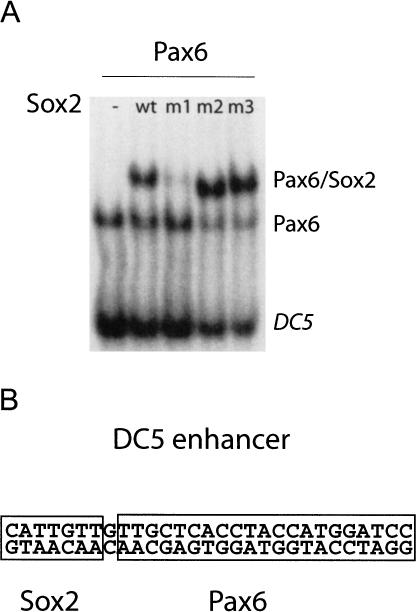
Sox2–Pax6 on DC5 and Sox2–Oct4 on FGF4 use the same HMG interface. (A) EMSA shows that the same mutation in HMG (m1) that interferes with POU/Sox2 on FGF4 also impairs complex formation with Pax6 on DC5, indicating that Sox2 uses the same interface for ternary complex assembly on these elements. (B) Schematic representation of the Sox2 and Pax6 DNA-binding site in the DC5 enhancer.
Sox2interacts with Oct4 and Pax6 via the same interface
SOX proteins can also establish direct functional partnerships with members of the PAX transcription factor family (Kamachi et al. 2000). PAX factors contain a conserved 128-amino-acid DNA-binding “paired domain” and play critical roles in mammalian development and oncogenesis (Mansouri et al. 1996). Sox2 specifically interacts with Pax6 in binding to the DC5 enhancer (Kamachi et al. 2001). This enhancer controls the expression of the δ-crystallin gene, which plays a pivotal role in eye developmentduring late embryogenesis (Kamachi et al. 2001).
To test whether Pax6 interacts with one of the two interfaces on Sox2, an EMSA with the DC5 elementwas performed with mutants m1, m2, and m3 in comparison to the wild-type HMG (Fig. 5). This experiment revealed that the same HMG domain mutation, R75E (m1), that specifically interfered with Oct/Sox2/FGF4 binding also abrogated Pax6/Sox2/DC5 ternary complex formation, whereas the UTF1-specific mutants (m2, m3) still interacted with Pax6. This finding suggests that the same Sox2 interface (the C-terminal region of HMG) is required for heterodimer formation with Oct4 on FGF4 and with Pax6 on DC5, although these two Sox2 partners are members of different transcription factor families and, as such, both unrelated in sequence and structure.
Discussion
POU and SOX proteins establish combinatorial developmental codes during development
Sox genes are expressed in various phases of embryonic development and cell differentiation. They are recognized as key players in the determination of cell fate (Pevny and Lovell-Badge 1997). Because HMG domains of SOX proteins are similar to each other in their DNA sequence preference (Mertin et al. 1999) and in their DNA-bending activity (Kamachi et al. 1999), it remains elusive how they are capable of specific target site selection. Assembly with unrelated transcriptional regulator proteins, however, provides a plausible explanation of how they can distinguish their targets as well as act in a cell-specific fashion (Kamachi et al. 2000). Partnering with members of the POU and PAX family of transcription factors may provide paradigm examples.
Interactions between various SOX and POU factors provide the best characterized examples for SOX partnership with members of another transcription factor family. There is a substantial number of well-characterized examples for this alliance in mouse (Oct-4/Sox-2, Oct-6/Sox-10, Brn-1,2/Sox-11) and in the fruit fly (Drifter/Dichaete) underlying their versatile involvement in different biological functions. Oct4/Sox2 partnership, for example, plays a fundamental role in determining the pluripotent cell state in early embryos (Yuan et al. 1995; Nishimoto et al. 1999). In contrast, Oct-6/Sox-10 and Brn-1,2/Sox-11 pairs have been shown to be involved in glial cell developmentin mouse (Kuhlbrodt et al. 1998). Another example is Drifter/Dichaete, which induces development of the central nervous system in the fruit fly (Soriano and Russell 1998). This latter finding might sug gest that POU/HMG partnership is an evolutionarily conserved developmental cue from invertebrates to mammals.
The list of SOX-interacting partners, however, is not limited to the POU and PAX families of transcription factors. Sox9, for example, activates its target gene Col2a1, which encodes type II collagen, during chondrogenesis in one case, and the anti-Müllerian (AMH) hormone gene during male sex determination in the genital ridge in another (Kamachi et al. 2000). In both cases, however, the proper expression pattern of the target gene requires the proximity of a conserved DNA-binding motif to the SOX-binding site on the regulator sequence as well as the concomitant binding of an additional factor. In the case of the AMH gene, this factor has been identified to be a member of the orphan nuclear receptor family (De Santa Barbara et al. 1998).
The crystal structure of the POU/HMG/DNA complex shed light onto the molecular mechanism of POU and SOX partnering on certain enhancers. It will be interesting to see if the principles unraveled in this work can be directly applied to other POU and SOX factor pairs or even—similarly to the resemblance of POU/SOX and PAX/SOX interaction—for partnership with various other protein families.
In vivo importance of differential POU/HMG interaction on FGF4 and UTF1
As FGF4 and UTF1 are differentially expressed during early mouse development, insight might be gained by comparing the activity of these two genes to the levels of their regulators: Sox2 and Oct4. The cell lines ES, F9 EC, and P19 EC are the in vitro counterpart of early stem-cell types of different embryonal stages and as such provide useful cell culture models (Yeom et al. 1996). Oct4 protein levels are similar in the three cell lines, whereas Sox2 protein levels vary (Table 2; Yeom et al. 1996; Botquin et al. 1998). An interesting finding is that FGF4 expression levels also vary in these cell lines (reverberating the pattern for Sox2), whereas UTF1 levels are similar (as are Oct4 levels; Table 2).
Table 2.
Expression levels of gene products in three different cell lines representing different cell types during early embryonic development
| ES cells | F9 EC cells | P19 EC cells | |
|---|---|---|---|
| Oct 4 | =a | =a | =a |
| Sox2 | Highb | Mediumb,c | Lowc |
| FGF4 | Highd | Mediumd | Lowd |
| UTF1 | =e | =e | =e |
= indicates that the level of a gene product is similar between the cell lines. The comparison is based on the following references:
Oct 4 (aBotquin et al. 1998); Sox2 (bDailey et al. 1994; cYuan et al. 1995); FGF4 (dSchoorlemmer and Kruijer 1991); and UTF1 (eOkuda et al. 1998).
One formidable hypothesis is that the different FGF4 and UTF1 activities during development are related to differences in the cooperativity of POU and HMG domain interactions on their respective enhancers. Differential cooperativity may provide a rationale for how the FGF4 and UTF1 genes respond to varying amounts of Oct4 and Sox2 proteins present during early development. Owing to a higher level of cooperativity, Oct4 may require less Sox2 to heterodimerize and augment UTF1 activity than is the case for FGF4. The differentdegrees of Sox2/Oct4 cooperativity on the regulatory elements in vitro are in congruence with the sequential up- and down-regulation of UTF1 and FGF4 during development, but these associations need to be tested in vivo. A stringent-functional test would require specific pointmutations to be introduced into the Oct4 and Sox2 genes, an approach that has become practical only after the construction of reliable three-dimensional models presented in this study.
Comparison of POU/POU homo- and POU/HMG heterodimerization
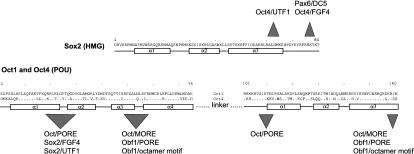
In earlier studies, we have reported that Oct factors are capable of homodimerization on two functionally distinct enhancers, termed PORE and MORE, and this binding was mediated by separate dimerization surface patches of their conserved POU domain (Fig. 6; Tomilin et al. 2000; Reményi et al. 2001b). Interestingly, Sox2 also contains two functionally and structurally distinct protein interaction surfaces. As is the case for previously mentioned POU factor dimers, the distance between the domain binding sites within the DNA motifs is critical for selecting between different interfaces of Sox2. This property is very likely to be instrumental in creating various multiprotein–DNA complexes with distinct biochemical properties. The differences in vitro, such as varying amount of cooperativity in complex formation, could result in distinct functional properties in vivo, such as varying amountof transcriptlevel production. Furthermore, the different quaternary arrangement of transcription factor/DNA complexes could also serve as the basis for differential recruitment of specific coregulators, as it has been shown for POU factor homodimerization of Oct1 and Pit1 on different enhancers (Scully et al. 2000; Tomilin et al. 2000; Reményi et al. 2001b, 2002).
Figure 6.
DNA-sequence-dependent protein interaction surfaces of Sox2 and Oct factors. Interface areas involved in protein—protein interaction in a DNA-sequence-specific manner are marked on the sequence of Sox2, Oct1, and Oct4 (nonvariant residues of Oct4 and Oct1 are shown with a dot). Several of these areas interact with several other protein partners, resulting in various combinations of homo- and heterodimers. The loop region between the first and second helices in the POU domain, for example, is involved in interactions with Sox2 on the FGF4 and UTF1 enhancers as well as in the homodimerization with a second Oct factor on the PORE element. Also, the C-terminal region the POU domain interacts not only with a second Oct factor on the MORE but, in the case of Oct1, also interacts with coactivator (Obf1) on the PORE as well as on the octamer motif(Chasman et al. 1999; Reményi et al. 2001b).
DNA-mediated interaction surface swapping as a general model
Our data support the emergence of a novel integrative approach to define the principles underlying differential complex formation onto DNA. Specifically, it subsumes that various combinations of transcription factors and their coregulators are possible, along with the potential of some of these proteins to interchange their quaternary DNA-mediated arrangements via one of multiple dimeric surfaces capable of protein–protein interactions (Fig. 6). Therefore, a certain dimerization surface patch appears to be adept at mediating glue-like surface interactions with different interacting protein partners in a versatile fashion. As such, our study provides insight into the adaptive mechanisms used by a finite set of transcription factors to assume a regulatory stronghold on various complex processes during mammalian development.
Materials and methods
Crystallization and structure determination
The POU domain (1–160 amino acids) of Oct1 and the HMG domain (1–80) of Sox2 were expressed in Escherichia coli and purified as described (Reményi et al. 2001a). The ternary complex was formed by mixing equivalentamounts of purified protein components with chemically synthesized DNA oligonucleotides (F22ta: 5′-TCTTTGTTTGGATGCTAATGGGa-3′ and F23ta: 5′-tTCTTTGTTTGGATGCTAATGGGA-3′) containing the natural sequence of the FGF4 enhancer element. POU/HMG:F22ta and POU/HMG:F23ta crystals suitable for X-ray analysis were grown with gel-filtrated ternary complex samples containing 12 mg/mL protein and 1.5-fold excess DNA at 20°C by using the hanging-drop vapor diffusion method. The POU/HMG:F22ta crystals were obtained from 35% (v/v) PEG550MME, 50 mM Na-citrate (pH 5.3); they belong to space group P6(4)22, with cell dimensions a = 119.2 Å, c = 154.5 Å. The POU/HMG:F23ta complex crystallized from 18% (v/w) PEG3350, 50 mM HEPES (pH 7.0), 20 mM MgCl2, and 5% (v/v) glycerol; this crystal form belongs to space group P3(1)21 with unitcell dimensions a = 72.8 Å, c = 172.4 Å.
The crystals were soaked briefly in a cryosolution containing mother liquor and 15% (v/v) glycerol, prior to mounting on nylon loops and flash-freezing in a nitrogen stream at 100 K. A native data set for the POU/HMG:F23ta complex crystals was collected to 2.6 Å resolution, using synchrotron radiation from Beamline BW7B at EMBL/DESY, Hamburg. Crystals of the POU/HMG:F22ta complex could also be obtained with a bromine derivative in which five thymine bases of the DNA oligonucleotide were replaced by 5-bromo-uracils (U): 5′-TCTUT GTUTGGAUGCTAATGGGa-3′, 5′-CCCAUTAGCAUCCAAA CAAAGAt-3′. A MAD data set at three wavelengths to 3.0 Å resolution was collected on a crystal at the BW7A Beamline at EMBL/DESY, Hamburg. Raw data were reduced and scaled with the HKL suite (Otwinowski and Minor 1997).
The structure of the POU/HMG:F22ta complex was solved by the MAD method (Table 1). The heavy-atom sites were identified using SHELX (Sheldrick 1998); their positions were refined and the phases were calculated in SHARP (La Fortelle and Bricogne 1997) with X-ray data between 20.0 and 3.0 Å resolution. The initial phases were further improved by solvent flattening with the SOLOMON of SHARP. An electron density map extending to 3.0 Å resolution was used for map interpretation and for manual model building in O (Jones et al. 1991). The ternary complex crystal structure was refined by using CNS (Brunger et al. 1998) to an R factor of 25%. This partially refined structural model was then used as a searching model in AMoRe (Navaza 1994) to solve the crystal structure of the better diffracting POU/HMG:F23ta complex. The 2.6-Å data set of this native complex was used for refinement of the structural model. The final model has an R factor of 23.2% (Rfree = 28.5%) and consists of the POU domain of Oct1 (POUS: 3–77 and POUH: 97–158), the HMG domain of Sox2 (1–79), and a 23-bp DNA oligonucleotide from the FGF4 enhancer. Although the visible part of the POUS–POUH linker is more extended than in earlier solved Oct1/DNA crystal structures (Klemm et al. 1994; Reményi et al. 2001b), the major part of this region still remains invisible.
Electrophoretic mobility shift assay (EMSA)
Wild-type and mutant versions of the HMG domain of Sox2 (K57E, R60E; R60E, M64E; and R75E) as well as the POU domain of Oct1 were expressed in E. coli with a histidine tag and purified with Ni-NTA agarose. Full-length Oct4, Oct4 I21Y,D29R, and a truncated version of Pax6 protein containing the Paired domain (1–169) were also expressed in E. coli and purified as described (Kamachi et al. 2001). Oligonucleotides containing natural sequences of the FGF4, UTF1, and DC5 enhancers were labeled with radioactivity: FGF4: ctgaAAGAAAACTCTTTGTTTGGATGCTAATGGGATACTAA Gctga; UTF1: ctgaAAGATGAGAGCCCTCATTGTTATGCTAGTGAAGTGCCAAGctga; DC5: ctgaTATTCATTGTTGTTGCTCACCTACCATGGATCCGAActga. (The POU, SOX and PAX binding sites are underlined.) Combinations of ∼25 ng of POU1, 100 ng of Oct4, 100 ng of Pax6, and 50 ng of HMG were incubated with the labeled oligonucleotides in the protein– DNA binding buffer (10 mM Tris atpH 8.0, 150 mM NaCl, 0.05% Triton-X, 5 μg/μL BSA, 50 ng/μL salmon testes DNA, 10% glycerol, 10 mM DTT), and complex formation was analyzed by EMSA. Titration experiments were carried out under similar conditions using the same amount Oct4 (100 ng) with increasing amounts of the HMG domain of Sox2 (0, 6.25, 12.5, 25, 50, and 100 ng) per binding reaction.
Acknowledgments
We are grateful to Ehmke Pohl and Peijian Zou for their practical advice. We thank H. Kondoh (University of Osaka, Japan) for providing plasmid for Pax6 expression. This work was started at the EMBL in Heidelberg, and finished at the EMBL in Hamburg and the New Bolton Center at the University of Pennsylvania. A.R is grateful to the EMBL for providing him with a short-term EMBL Postdoctoral Fellowship. This work was supported in partby HFSP grantRGP 62/2002 (H.R.S. and M.W.), the Marion Dilley and David George Jones Funds, and the Commonwealth and General Assembly of Pennsylvania (H.R.S.). Coordinates of the Oct1/Sox2/FGF4 ternary complex have been deposited in the Protein Data Bank under accession code 1gt0.
The publication costs of this article were defrayed in part by paymentof page charges. This article musttherefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.269303.
Supplemental material is available online at http://www.genesdev.org.
References
- Ambrosetti, D.C., Basilico, C., and Dailey, L. 1997. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol. Cell. Biol. 17: 6321–6329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosetti, D.C., Scholer, H.R., Dailey, L., and Basilico, C. 2000. Modulation of the activity of multiple transcriptional activation domains by the DNA binding domains mediates the synergistic action of Sox2 and Oct-3 on the fibroblast growth factor-4 enhancer. J. Biol. Chem. 275: 23387–23397. [DOI] [PubMed] [Google Scholar]
- Avilion, A.A., Nicolis, S.K., Pevny, L.H., Perez, L., Vivian, N., and Lovell-Badge, R. 2003. Multipotent cell lineages in early mouse developmentdepend on SOX2 function. Genes & Dev. 17: 126–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botquin, V., Hess, H., Fuhrmann, G., Anastassiadis, C., Gross, M.K., Vriend, G., and Scholer, H.R. 1998. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes & Dev. 12: 2073–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Chasman, D., Cepek, K., Sharp, P.A., and Pabo, C.O. 1999. Crystal structure of an OCA-B peptide bound to an Oct-1 POU domain/octamer DNA complex: Specific recognition of a protein–DNA interface. Genes & Dev. 13: 2650–2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dailey, L. and Basilico, C. 2001. Coevolution of HMG domains and homeodomains and the generation of transcriptional regulation by Sox/POU complexes. J. Cell Physiol. 186: 315–328. [DOI] [PubMed] [Google Scholar]
- Dailey, L., Yuan, H., and Basilico, C. 1994. Interaction between a novel F9-specific factor and octamer-binding proteins is required for cell-type-restricted activity of the fibroblast growth factor 4 enhancer. Mol. Cell. Biol. 14: 7758–7769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa Barbara, P., Bonneaud, N., Boizet, B., Desclozeaux, M., Moniot, B., Sudbeck, P., Scherer, G., Poulat, F., and Berta, P. 1998. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol. Cell. Biol. 18: 6653–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herr, W. and Cleary, M.A. 1995. The POU domain: Versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes & Dev. 9: 1679–1693. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard. 1991. Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kamachi, Y., Cheah, K.S., and Kondoh, H. 1999. Mechanism of regulatory target selection by the SOX high-mobility-group domain proteins as revealed by comparison of SOX1/2/3 and SOX9. Mol. Cell. Biol. 19: 107–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamachi, Y., Uchikawa, M., and Kondoh, H. 2000. Pairing SOX off: With partners in the regulation of embryonic development. Trends Genet. 16: 182–187. [DOI] [PubMed] [Google Scholar]
- Kamachi, Y., Uchikawa, M., Tanouchi, A., Sekido, R., and Kondoh, H. 2001. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes & Dev. 15: 1272–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm, J.D., Rould, M.A., Aurora, R., Herr, W., and Pabo, C.O. 1994. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell 77: 21–32. [DOI] [PubMed] [Google Scholar]
- Kuhlbrodt, K., Herbarth, B., Sock, E., Enderich, J., Hermans-Borgmeyer, I., and Wegner, M. 1998. Cooperative function of POU proteins and SOX proteins in glial cells. J. Biol. Chem. 273: 16050–16057. [DOI] [PubMed] [Google Scholar]
- La Fortelle, E. and Bricogne, G. 1997. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD method. In Methods in enzymology, macromolecular crystallography (eds. R.M. Sweetand C.W.J. Carter), pp. 472–494. Academic Press, New York. [DOI] [PubMed]
- Lavery, R. and Sklenar, H. 1988. The definition of generalized helicoidal parameters and of axis curvature for irregular nucleic acids. J. Biomol. Struct. Dyn. 6: 63–91. [DOI] [PubMed] [Google Scholar]
- Love, J.J., Li, X., Case, D.A., Giese, K., Grosschedl, R., and Wright, P.E. 1995. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376: 791–795. [DOI] [PubMed] [Google Scholar]
- Mansouri, A., Hallonet, M., and Gruss, P. 1996. Pax genes and their roles in cell differentiation and development. Curr. Opin. Cell Biol. 8: 851–857. [DOI] [PubMed] [Google Scholar]
- Mertin, S., McDowall, S.G., and Harley, V.R. 1999. The DNA-binding specificity of SOX9 and other SOX proteins. Nucleic Acids Res. 27: 1359–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, F.V.T. and Churchill, M.E. 2000. Nonsequence-specific DNA recognition: A structural perspective. Structure Fold Des. 8: R83–R89. [DOI] [PubMed] [Google Scholar]
- Murphy, F.V.T., Sweet, R.M., and Churchill, M.E. 1999. The structure of a chromosomal high mobility group protein– DNA complex reveals sequence-neutral mechanisms important for non-sequence-specific DNA recognition. EMBO J. 18: 6610–6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza, J. 1994. AMoRe: An automated package for molecular replacement. Acta Crystallogr. A 50: 157–163. [Google Scholar]
- Nichols, J., Zevnik, B., Anastassiadis, K., Niwa, H., Klewe-Nebenius, D., Chambers, I., Scholer, H., and Smith, A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 95: 379–391. [DOI] [PubMed] [Google Scholar]
- Nishimoto, M., Fukushima, A., Okuda, A., and Muramatsu, M. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol. Cell. Biol. 19: 5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa, H., Miyazaki, J., and Smith, A.G. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 24: 372–376. [DOI] [PubMed] [Google Scholar]
- Okuda, A., Fukushima, A., Nishimoto, M., Orimo, A., Yamagishi, T., Nabeshima, Y., Kuro-o, M., Boon, K., Keaveney, M., Stunnenberg, H.G., et al. 1998. UTF1, a novel transcriptional coactivator expressed in pluripotent embryonic stem cells and extra-embryonic cells. EMBO J. 17: 2019–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymology A 276: 307–325. [DOI] [PubMed] [Google Scholar]
- Pevny, L.H. and Lovell-Badge, R. 1997. Sox genes find their feet. Curr. Opin. Genet. Dev. 7: 338–344. [DOI] [PubMed] [Google Scholar]
- Reményi, A., Pohl, E., Scholer, H.R., and Wilmanns, M. 2001a. Crystallization of redox-insensitive Oct1 POU domain with differentDNA-response elements. Acta Crystallogr. D Biol. Crystallogr. 57: 1634–1638. [DOI] [PubMed] [Google Scholar]
- Reményi, A., Tomilin, A., Pohl, E., Lins, K., Philippsen, A., Reinbold, R., Scholer, H.R., and Wilmanns, M. 2001b. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol. Cell 8: 569–580. [DOI] [PubMed] [Google Scholar]
- Reményi, A., Tomilin, A., Scholer, H.R., and Wilmanns, M. 2002. Differential activity by DNA-induced quarternary structures of POU transcription factors. Biochem. Pharmacol. 64: 979–984. [DOI] [PubMed] [Google Scholar]
- Schoorlemmer, J. and Kruijer, W. 1991. Octamer-dependent regulation of the kFGF gene in embryonal carcinoma and embryonic stem cells. Mech. Dev. 36: 75–86. [DOI] [PubMed] [Google Scholar]
- Scully, K.M., Jacobson, E.M., Jepsen, K., Lunyak, V., Viadiu, H., Carriere, C., Rose, D.W., Hooshmand, F., Aggarwal, A.K., and Rosenfeld, M.G. 2000. Allosteric effects of Pit-1 DNA sites on long-term repression in cell type specification. Science 290: 1127–1131. [DOI] [PubMed] [Google Scholar]
- Sheldrick, G.M. 1998. SHELX: Application to macromolecules. In Direct methods for solving macromolecular structures (ed. S. Fortier), pp. 401–411. Kluwer Academic Publishers, Dordrecht.
- Soriano, N.S. and Russell, S. 1998. The Drosophila SOX-domain protein Dichaete is required for the development of the central nervous system midline. Development 125: 3989–3996. [DOI] [PubMed] [Google Scholar]
- Tomilin, A., Reményi, A., Lins, K., Bak, H., Leidel, S., Vriend, G., Wilmanns, M., and Scholer, H.R. 2000. Synergism with the coactivator OBF-1 (OCA-B, BOB-1) is mediated by a specific POU dimer configuration. Cell 103: 853–864. [DOI] [PubMed] [Google Scholar]
- Vriend, G. 1990. WHATIF: A molecular modeling and drug design program. J. Mol. Graph. 8: 52–56. [DOI] [PubMed] [Google Scholar]
- Wegner, M. 1999. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res. 27: 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner, M.H., Huth, J.R., Gronenborn, A.M., and Clore, G.M. 1995. Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY–DNA complex. Cell 81: 705–714. [DOI] [PubMed] [Google Scholar]
- Yeom, Y.I., Fuhrmann, G., Ovitt, C.E., Brehm, A., Ohbo, K., Gross, M., Hubner, K., and Scholer, H.R. 1996. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development 122: 881–894. [DOI] [PubMed] [Google Scholar]
- Yuan, H., Corbi, N., Basilico, C., and Dailey, L. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes & Dev. 9: 2635–2645. [DOI] [PubMed] [Google Scholar]