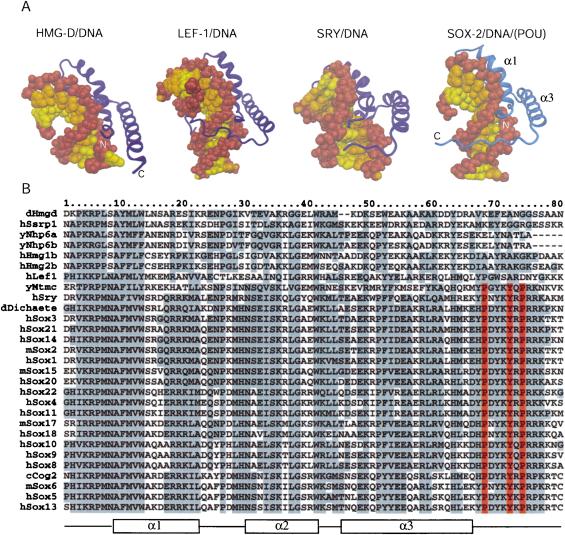
Figure 3.
Comparison of differentHMG domains. (A) Structures of different HMG domains from HMG-D (Murphy et al. 1999), Lef-1 (Love et al. 1995), Sry (Werner et al. 1995), and Sox2 (taken from the Oct1/Sox2/FGF4 crystal structure) in complex with DNA. Sox2 generates a similar bend angle (90°) in DNA as reported for other HMG/DNA complex structures (HMG-D, 111°; Lef-1, 117°; Sry, 75°). Helix 1 and helix 2 form extensive contacts with the DNA in the widened minor groove and are involved in bending the DNA in all structures. However, the proteins show significant differences in how their C-terminal regions interact with the DNA molecule. In HMG-D, which binds DNA without sequence specificity, only helices 1 and 2 interact with the DNA. The C terminus of the Lef-1 HMG domain lies in the compressed major groove and stabilizes the bent DNA conformation. In the Sry/DNA structure, the C-terminal part is mainly disordered and is not positioned in the minor groove. The Sox2-HMG C-terminal region fits tightly into the compressed minor groove when in the presence of the POUS domain. The DNA sugar-phosphate backbone is brown, and the bases belonging to different chains of the DNA molecule are depicted by differentcolors (yellow and orange). Helix 2 (α2) is behind the DNA molecule and, therefore, cannot be seen from this orientation. (B) Multiple sequence alignment of HMG domains of several proteins from differentorganisms. (y) Yeast; (c) Caenorhabditis elegans; (d) Drosophila melanogaster; (m) mouse; (h) human. The order of the proteins on the list reflects their sequence similarity. HMG domains from the first six proteins (dHmgd–hHmg2b) are known to bind DNA in a non-sequence-specific manner, whereas the others (hLef1–hSox13) bind DNA according to a specific sequence. Secondary structure elements of Sox2 from the Oct1/Sox2/DNA crystal structure are shown beneath the alignment. Protein residues that are highly conserved are boxed in gray. It is notable that the protein residues that play an important role in the ordering of the C-terminal region of the HMG from Sox2 (V3, R5, P6, H63, H67, P68, Y70, Y72, R75, and R76) are conserved in almostall members of the SOX family. However, these residues are divergent in HMG domains that bind DNA in a non-sequence-specific manner (Murphy and Churchill 2000). Residues P68, Y72, and P74, which play the most prominent role in positioning the C terminus in the compressed minor groove (cf. Fig. 4A), are red.