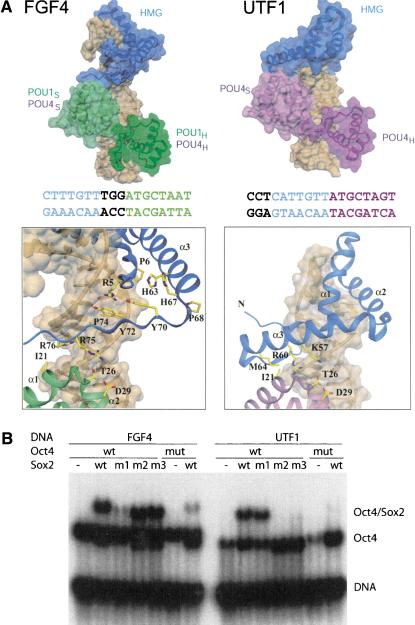
Figure 4.
Comparison of POU/HMG complexes formed on FGF4 and UTF1. (A, top) Model of POU/HMG/FGF4 (POU domain of Oct1 or Oct4; left), compared with the model of Oct4-POU/HMG/UTF1 (right). The figure illustrates that different spacing between the binding sites for the POU and HMG domains within FGF4 and UTF1 enhancers causes formation of different heterodimeric interfaces. (Bottom) Close-up views of the HMG/POUS interfaces on FGF4 (left) and UTF1 (right). The POU/HMG/UTF1 model suggests involvement of helix 3 instead of the C terminus of the HMG domain to form the HMG-POUS interface, whereas the same surface patch of the POUS domain appears to be involved in both interfaces. The DNA molecules are depicted with a transparent surface and are brown. Coloring of the FGF4 and UTF1 DNA sequences is according to their POU- and Sox2-HMG-binding sites. Notice the difference in spacing of these two sites in the two enhancers. (B) To validate the homology models in A, mutations in Sox2-HMG were designed to selectively interfere with ternary complex formation on the FGF4 (m1) or on the UTF1 enhancers (m2 and m3); (m1) R75E; (m2) K57E,R60E; (m3) R60E,M64E; (mut) mutant version of Oct4 POU (I21Y,D29E). In agreement with both models, m1 specifically impaired heterodimerization on FGF4, whereas m2 and m3 specifically abrogated heterodimerization on UTF1, but Oct4 mut affected both.