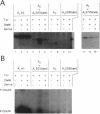
Abstract
Escherichia coli cells express two forms of the chemotaxis-associated CheA protein, CheAL and CheAS, as the result of translational initiation at two distinct in-frame initiation sites in the gene cheA. The long form, CheAL, plays a crucial role in chemotactic signal transduction. As a histidine protein kinase, it first autophosphorylates at amino acid His-48; then, it phosphorylates two other chemotaxis proteins, CheY and CheB. The short form, CheAS, lacks the amino-terminal 97 amino acids of CheAL and, therefore, does not contain the site of autophosphorylation. However, it does retain a functional kinase domain. As a consequence, CheAS can mediate transphosphorylation of kinase-deficient CheAL variants. Here we demonstrate in vitro that CheAS also can mediate transphosphorylation of a CheAL variant that lacks the C-terminal segment, a portion of the protein which is thought to interact with CheW and the chemoreceptors. The presence of CheW and the chemoreceptor Tsr enhances this activity and results in modulation of the transphosphorylation rate in response to the Tsr ligand, L-serine. Because CheAS can mediate this activity, it can restore chemotactic ability to Escherichia coli cells that express this truncated CheAL variant.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adler J. Chemotaxis in bacteria. Science. 1966 Aug 12;153(3737):708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- Bennett M. J., Choe S., Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci U S A. 1994 Apr 12;91(8):3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair D. F., Berg H. C. Restoration of torque in defective flagellar motors. Science. 1988 Dec 23;242(4886):1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Alex L. A., Simon M. I. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci U S A. 1992 Aug 1;89(15):6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Kaplan N., Hess J. F., Simon M. I. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc Natl Acad Sci U S A. 1989 Feb;86(4):1208–1212. doi: 10.1073/pnas.86.4.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkovich K. A., Simon M. I. Coupling of receptor function to phosphate-transfer reactions in bacterial chemotaxis. Methods Enzymol. 1991;200:205–214. doi: 10.1016/0076-6879(91)00140-r. [DOI] [PubMed] [Google Scholar]
- Borkovich K. A., Simon M. I. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell. 1990 Dec 21;63(6):1339–1348. doi: 10.1016/0092-8674(90)90429-i. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Borkovich K. A., Simon M. I. Signal transduction pathways involving protein phosphorylation in prokaryotes. Annu Rev Biochem. 1991;60:401–441. doi: 10.1146/annurev.bi.60.070191.002153. [DOI] [PubMed] [Google Scholar]
- Bourret R. B., Davagnino J., Simon M. I. The carboxy-terminal portion of the CheA kinase mediates regulation of autophosphorylation by transducer and CheW. J Bacteriol. 1993 Apr;175(7):2097–2101. doi: 10.1128/jb.175.7.2097-2101.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner J. A., Dahlquist F. W. Signal transduction in bacteria: CheW forms a reversible complex with the protein kinase CheA. Proc Natl Acad Sci U S A. 1991 Feb 1;88(3):750–754. doi: 10.1073/pnas.88.3.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegner J. A., Graham D. R., Roth A. F., Dahlquist F. W. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992 Sep 18;70(6):975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- Gill S. C., von Hippel P. H. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989 Nov 1;182(2):319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Bourret R. B., Simon M. I. Histidine phosphorylation and phosphoryl group transfer in bacterial chemotaxis. Nature. 1988 Nov 10;336(6195):139–143. doi: 10.1038/336139a0. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Kaplan N., Simon M. I. Phosphorylation of three proteins in the signaling pathway of bacterial chemotaxis. Cell. 1988 Apr 8;53(1):79–87. doi: 10.1016/0092-8674(88)90489-8. [DOI] [PubMed] [Google Scholar]
- Hess J. F., Oosawa K., Matsumura P., Simon M. I. Protein phosphorylation is involved in bacterial chemotaxis. Proc Natl Acad Sci U S A. 1987 Nov;84(21):7609–7613. doi: 10.1073/pnas.84.21.7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofoid E. C., Parkinson J. S. Tandem translation starts in the cheA locus of Escherichia coli. J Bacteriol. 1991 Mar;173(6):2116–2119. doi: 10.1128/jb.173.6.2116-2119.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krikos A., Conley M. P., Boyd A., Berg H. C., Simon M. I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci U S A. 1985 Mar;82(5):1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Genetic evidence for interaction between the CheW and Tsr proteins during chemoreceptor signaling by Escherichia coli. J Bacteriol. 1991 Aug;173(16):4941–4951. doi: 10.1128/jb.173.16.4941-4951.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. D., Parkinson J. S. Role of CheW protein in coupling membrane receptors to the intracellular signaling system of bacterial chemotaxis. Proc Natl Acad Sci U S A. 1989 Nov;86(22):8703–8707. doi: 10.1073/pnas.86.22.8703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura P., Rydel J. J., Linzmeier R., Vacante D. Overexpression and sequence of the Escherichia coli cheY gene and biochemical activities of the CheY protein. J Bacteriol. 1984 Oct;160(1):36–41. doi: 10.1128/jb.160.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally D. F., Matsumura P. Bacterial chemotaxis signaling complexes: formation of a CheA/CheW complex enhances autophosphorylation and affinity for CheY. Proc Natl Acad Sci U S A. 1991 Jul 15;88(14):6269–6273. doi: 10.1073/pnas.88.14.6269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison T. B., Parkinson J. S. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci U S A. 1994 Jun 7;91(12):5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa E. G., Atkinson M. R., Kamberov E. S., Ninfa A. J. Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits. J Bacteriol. 1993 Nov;175(21):7024–7032. doi: 10.1128/jb.175.21.7024-7032.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninfa E. G., Stock A., Mowbray S., Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991 May 25;266(15):9764–9770. [PubMed] [Google Scholar]
- Oosawa K., Hess J. F., Simon M. I. Mutants defective in bacterial chemotaxis show modified protein phosphorylation. Cell. 1988 Apr 8;53(1):89–96. doi: 10.1016/0092-8674(88)90490-4. [DOI] [PubMed] [Google Scholar]
- Park C., Hazelbauer G. L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986 Jul;167(1):101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Houts S. E. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J Bacteriol. 1982 Jul;151(1):106–113. doi: 10.1128/jb.151.1.106-113.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J. S., Kofoid E. C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Smith R. A., Parkinson J. S. Overlapping genes at the cheA locus of Escherichia coli. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5370–5374. doi: 10.1073/pnas.77.9.5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stader J., Matsumura P., Vacante D., Dean G. E., Macnab R. M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986 Apr;166(1):244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A., Chen T., Welsh D., Stock J. CheA protein, a central regulator of bacterial chemotaxis, belongs to a family of proteins that control gene expression in response to changing environmental conditions. Proc Natl Acad Sci U S A. 1988 Mar;85(5):1403–1407. doi: 10.1073/pnas.85.5.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock A., Koshland D. E., Jr, Stock J. Homologies between the Salmonella typhimurium CheY protein and proteins involved in the regulation of chemotaxis, membrane protein synthesis, and sporulation. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7989–7993. doi: 10.1073/pnas.82.23.7989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock J. B., Lukat G. S., Stock A. M. Bacterial chemotaxis and the molecular logic of intracellular signal transduction networks. Annu Rev Biophys Biophys Chem. 1991;20:109–136. doi: 10.1146/annurev.bb.20.060191.000545. [DOI] [PubMed] [Google Scholar]
- Swanson R. V., Bourret R. B., Simon M. I. Intermolecular complementation of the kinase activity of CheA. Mol Microbiol. 1993 May;8(3):435–441. doi: 10.1111/j.1365-2958.1993.tb01588.x. [DOI] [PubMed] [Google Scholar]
- Swanson R. V., Schuster S. C., Simon M. I. Expression of CheA fragments which define domains encoding kinase, phosphotransfer, and CheY binding activities. Biochemistry. 1993 Aug 3;32(30):7623–7629. doi: 10.1021/bi00081a004. [DOI] [PubMed] [Google Scholar]
- Wolfe A. J., Berg H. C. Migration of bacteria in semisolid agar. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe A. J., Stewart R. C. The short form of the CheA protein restores kinase activity and chemotactic ability to kinase-deficient mutants. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1518–1522. doi: 10.1073/pnas.90.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton J. C., Drummond M. H. The Q-linker: a class of interdomain sequences found in bacterial multidomain regulatory proteins. Protein Eng. 1989 May;2(7):535–543. doi: 10.1093/protein/2.7.535. [DOI] [PubMed] [Google Scholar]
- Yang Y., Inouye M. Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli. Proc Natl Acad Sci U S A. 1991 Dec 15;88(24):11057–11061. doi: 10.1073/pnas.88.24.11057. [DOI] [PMC free article] [PubMed] [Google Scholar]