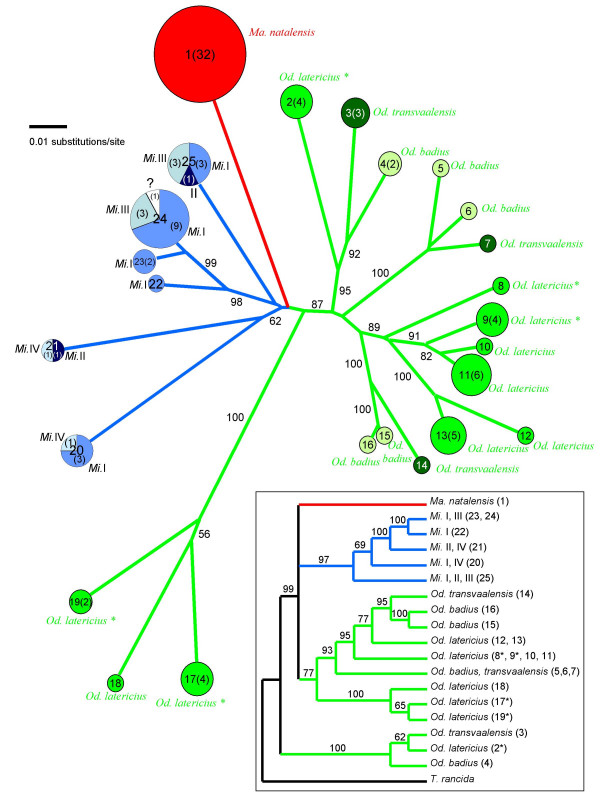
Figure 2.
a. Unrooted neighbor-joining phylogram of the 25 ITS haplotypes found among the Termitomyces symbionts of 101 nests of fungus growing termites (belonging to eight species in three genera). The three genera are indicated with different colours, and the species within the genera in different intensities of these colours. The Senegalese samples of O. latericius are indicated with an asterisk. Within each circle the haplotype number is indicated and, in brackets, the number of that haplotype found (if more than one). The area of circles is proportional to the observed frequency of particular haplotypes. Red: genus Macrotermes; green: genus Odontotermes; blue: genus Microtermes. Within the genus Microtermes four tentative species were distinguished (labelled I-IV). For one Microtermes termite sample no sequence was obtained, which is indicated with a '?'. The numbers on the branches are the percentage bootstrap values (> 50) based on 1000 bootstrap replicates. b. Phylogenetic relationships between the Termitomyces symbionts of the fungus growing termites included in this study. The cladogram is the majority rule consensus tree of trees sampled in a Bayesian analysis of combined partial nuclear 25S sequences and mitochondrial 12S sequences. The free-living fungus Tephrocybe rancida was used as an outgroup, based on previous analyses (Hoffstetter et al. 2002; Aanen et al., 2002). Abbreviations used: Ma.: Macrotermes; Mi.: Microtermes; Od.: Odontotermes.