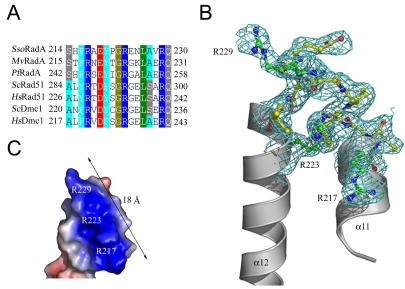
Figure 2. Structure of the L1 motif.
(A) Sequence alignment of RadA homologs from S. solfataricus (Sso) RadA, M. voltae (Mv) RadA, P. furiosus (Pf) Rad51, H. sapiens (Hs) Rad51 and Dmc1, and S. cerevisiae (Sc) Rad51 and Dmc1. Three conserved arginine residues are shown in cyan. (B) A ribbon diagram of the L1 motif showing two alpha-helices (grey). The hinge region is depicted with a ball-and-stick model (yellow). The side chains of three conserved arginine residues are shown in green. 2F o–F c electron density maps (contoured at 1.0σ), corresponding to the ssDNA binging site, are shown in cyan. (C) Surface charge potential of the L1 motif. The positively and negatively charged regions are indicated in blue and red, respectively. The linear basic patch is ∼18 Å in length.