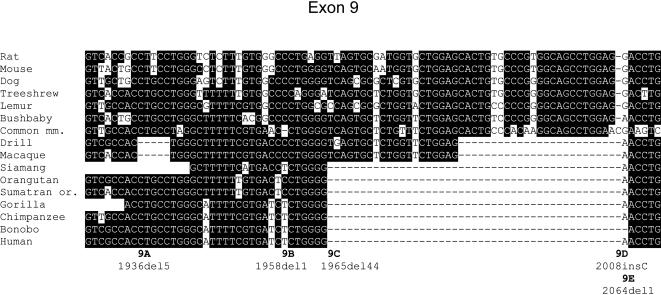
Figure 2. Representative GC-D exon alignment showing multiple inactivating mutations.
Alignment of GC-D exon 9 sequences from rat, mouse, dog, treeshrew and various primates. At each alignment position, sequences matching the consensus are shown as white letters with black background, and sequences that do not match are black on white. Insertions/deletions are shown as “−“ characters; areas of missing sequence are entirely blank. Each inactivating mutation is labeled below the alignment: e.g. 9A is the 5′-most mutation in exon 9. Note that the 24-bp deletion in drill and macaque does not introduce a frameshift: it is thus unclear whether it would interfere with function. A full alignment is provided in Figure S1. Species abbreviations: mm., marmoset; or., orangutan.