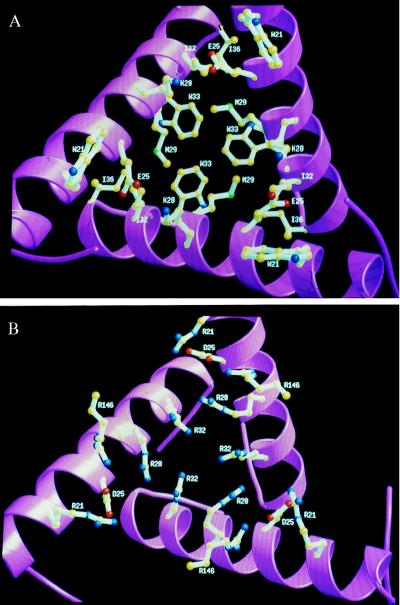
Figure 2.
View of the interface between trimers located around a 3-fold symmetry axis at one top of the dodecamer. The three H1 helices from CP domains that belong to three monomers are shown in ribbons. (A) Interface in PF, mainly composed of the hydrophobic residues W21, M29, I32, W33, and I36. The salt bridges between K28 and E25 also are shown. (B) Interface in PA composed of positively charged residues: R21, R28, R32, and R146, constituting a binding site for negatively charged allosteric effectors. The difference in compactness between both enzymes also is illustrated by the increased packing in the PA OTCase of the three helices H1 around the 3-fold symmetry axis. Figures were generated by using molscript (36) and raster3d (37).