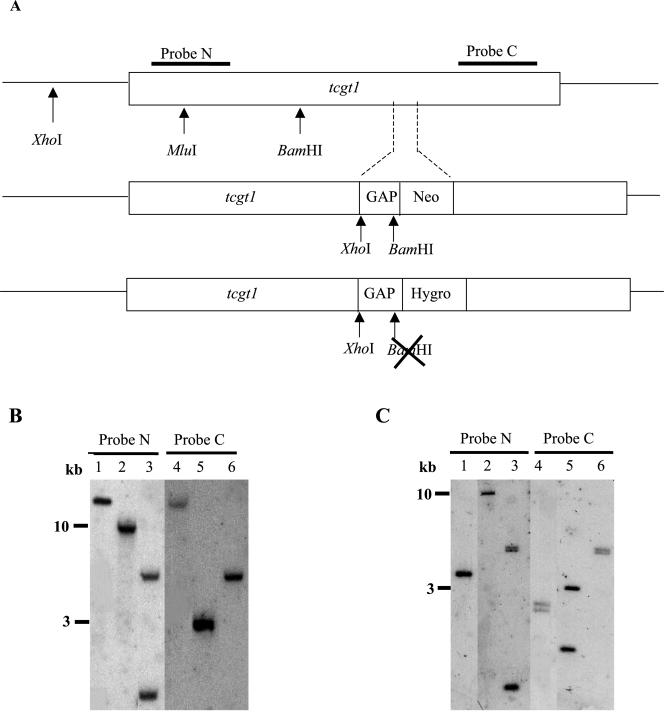
Figure 2.
Characterization of tcgt1 copy number and tcgt1–/tcgt1– (GT null) mutants. Restriction enzyme cleavage sites in tcgt1 and in the disrupted gene as well as coding regions of probes used are indicated in A. The BamHI site in GAP disappears upon cleaving the Neomycin-resistant construction with EcoRI. Wild-type (B) and tcgt1–/tcgt1– (GT null) mutant (C) cell DNAs were subjected to restriction enzyme cleavage followed by Southern blotting analysis using indicated probes. Restriction enzymes used were XhoI (lanes 1 and 4), BamHI (lanes 2 and 5), and MluI (lanes 3 and 6). GAP, Neo, and Hygro stand for a 470-base pair fragment of the 5′ UTR of the glyceraldehydephosphate dehydrogenase encoding gene (GAP), containing the splice acceptor site, and genes conferring resistance to Neomycin and Hygromycin, respectively. For further details, see MATERIALS AND METHODS.