Figure 1.
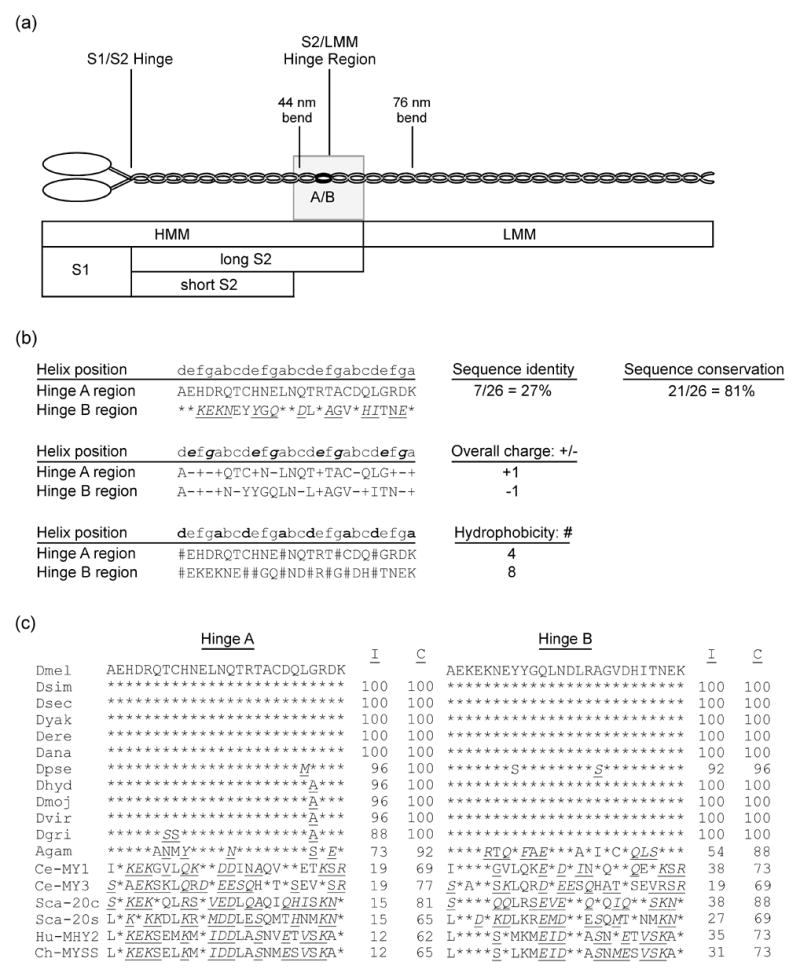
Common proteolytic sites of myosin, highlighting the S2/LMM hinge region, and analysis of its variant sequences in Drosophila and other organisms. (a) MHC molecules dimerize by intertwining of their alpha-helical coiled-coil tails. Each MHC consists of a globular head, an alpha-helical rod, and a non-helical C-terminal tailpiece. The essential and regulatory light chains are not shown. The MHC rod is ~155 nm long5,51,79 and can be proteolytically cleaved into heavy meromyosin (HMM) and light meromyosin (LMM). HMM can be further digested to generate subfragment 1 (S1), containing the head-neck region, as well as (long) subfragment 2 (S2). S2 can be further digested into short S2 and the C-terminal S2 hinge region also designated as the S2/LMM hinge (shaded rectangle). Bends in the rod are observed at 44 nm and 76 nm from the head-rod junction.5 The black oval labeled A/B indicates the approximate location and relative size of the 26-residue region encoded by alternative exons 15a and 15b in Drosophila muscle myosin. Scale is approximate. (b) Comparison of sequence, charge and hydrophobic variation of hinge A with hinge B. Lower case letters indicate amino acid residue positions in the alpha-helical heptad repeat.1 Top: * indicates identical residues; underline indicates conserved residues [italic type: conservation of strong groups (suggesting both hydropathy and side group size conservation), roman type: conservation of weak groups (suggesting either hydropathy or side group size conservation)]. Middle: bold italic indicates positions e and g, which are typically occupied by charged residues.1 (+) or (−) indicates charged residues in hinges A and B. Bottom: bold indicates positions a and d, which typically contain hydrophobic residues.1 # indicates hydrophobic residues in hinges A and B. (c) Sequence comparison of hinges A and B of D. melanogaster with those of muscle myosin from other Drosophila species, mosquito, worm, bay scallop, chicken and human. * indicates identical residues; underline indicates conserved residues (italic type: conservation of strong groups, roman type: conservation of weak groups). I, % identical residues, C, % conserved residues (percentage of residues that are identical, or show strong or weak conservation with the D. melanogaster hinge sequence below which they are listed). Abbreviations: Dmel, D. melanogaster; Dsim, D. simulans; Dsec, D. sechellia; Dyak; D. yakuba; Dere; D. erecta; Dana, D. ananassae; Dpse, D. pseudoobscura; Dhyd, D. hydei; Dmoj, D. mojavensis; Dvir, D. virulis; Dgri, D. grimshawi; Agam, Anopheles gambiae; Ce-MY1, Caenorhabditis elegans MHC D; Ce-MY3, C. elegans MHC A; Sca-20c, bay scallop, Aequipecten irradians, catch muscle; Sca-20s, A. irradians striated muscle; Hu-MHY2, human, MHC-IIa; Ch-MYSS, chicken adult skeletal muscle MHC.
