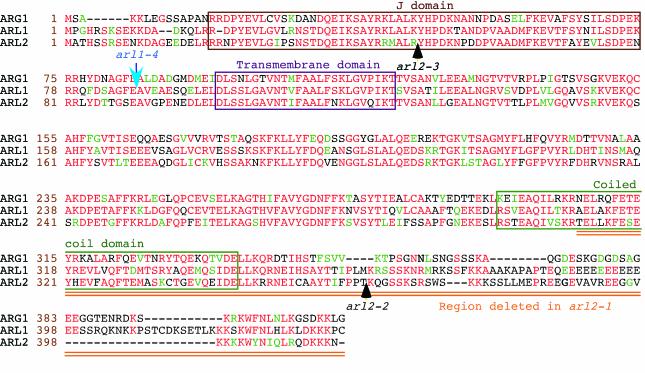
Figure 6.
Amino acid sequence alignments of the ARG1 (top), ARL1 (middle), and ARL2 (bottom) proteins (Ws ecotype). Amino acid positions within each protein are shown at the left of each sequence (brown numbers). Protein names are indicated at the left of these numbers. Amino acids that are identical or conserved between at least two of the three proteins are shown in red and green characters, respectively. Gaps introduced within a protein to allow better alignment of flanking sequences are represented by dashes. The J domain, putative transmembrane domain, and predicted coiled-coil region are boxed. The arl2-1 mutation deletes the doubly underlined (orange) carboxyl end of the protein, whereas positions of T-DNA inserts within ARL2 and ARL1 alleles are indicated by black and blue arrows, respectively. Alignments were obtained by the Boxshade server (Institut Pasteur, Paris; http://bioweb.pasteur.fr/seqanal/interfaces/boxshade.html).