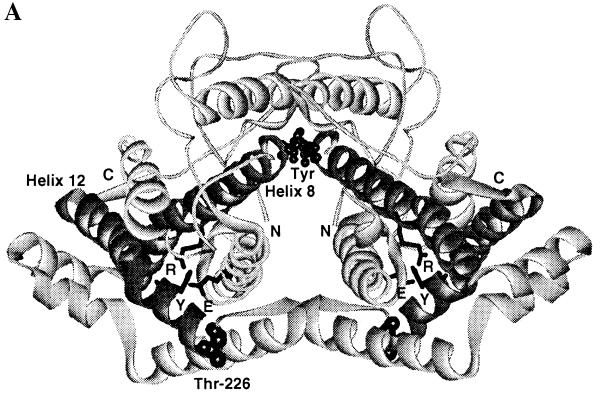
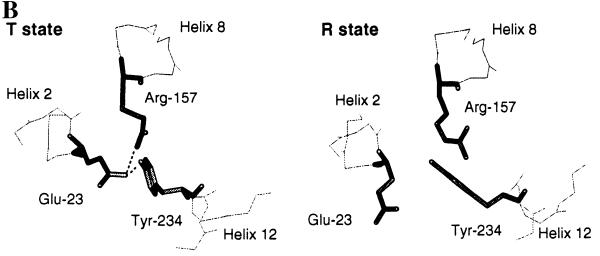
Figure 2.
(A) Structure of the T state of chorismate mutase drawn in ribbon style. The N termini and C termini are labeled as N and C, respectively. The two tyrosine molecules located at the interface of the dimer are indicated as Tyr. Helix 8, which connects the allosteric and active sites, and helix 12 are drawn in a dark color. The side chains of Glu-23 (E) and Tyr-234 (Y), which are involved in the hydrogen-bonding interaction with the active residue Arg-157 (R), are drawn in stick model. Thr-226, which connects the 220s loop and helix 12, is indicated. (B) Residues Glu-23, Tyr-234, and Arg-157 in the crystal structures of the R and T states of yeast chorismate mutase. Hydrogen-bonding interactions are indicated by dotted lines. Glu-23 and Tyr-234 move toward the active site upon the transition from the R to the T state. In the T state, Glu-23 is in hydrogen-bonding distance to the active site residue Arg-157, and the OH of Tyr-234 is 2.7 Å from Oɛ2 of Glu-23; in the R state this distance increases to 4.7 Å.