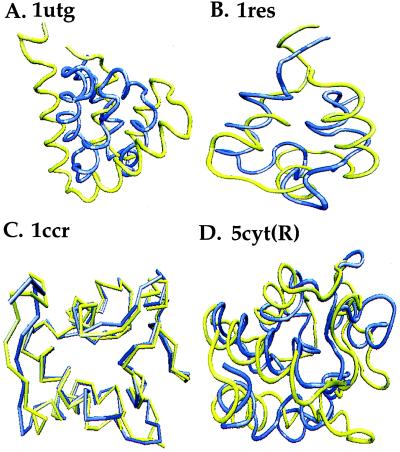
Figure 5.
Superposition of the predicted (blue) and native (yellow) structures of (A) uteroglobin (1utg), (B) gamma delta resolvase (1res), (C) rice embryo cytochrome c (1ccr), and (D) cytochrome c (5cyt(R)). All of the predicted structures show the correct topology for the native structure although each predicted structure tends to be slightly overcollapsed. The 1utg predicted structure has the largest deviation from its native structure caused by overcollapse. 1res is one of the two predicted structures that exhibit “creativity,” i.e., the rms of the predicted structure is higher than the rms of the best scaffold in its memory set.