Abstract
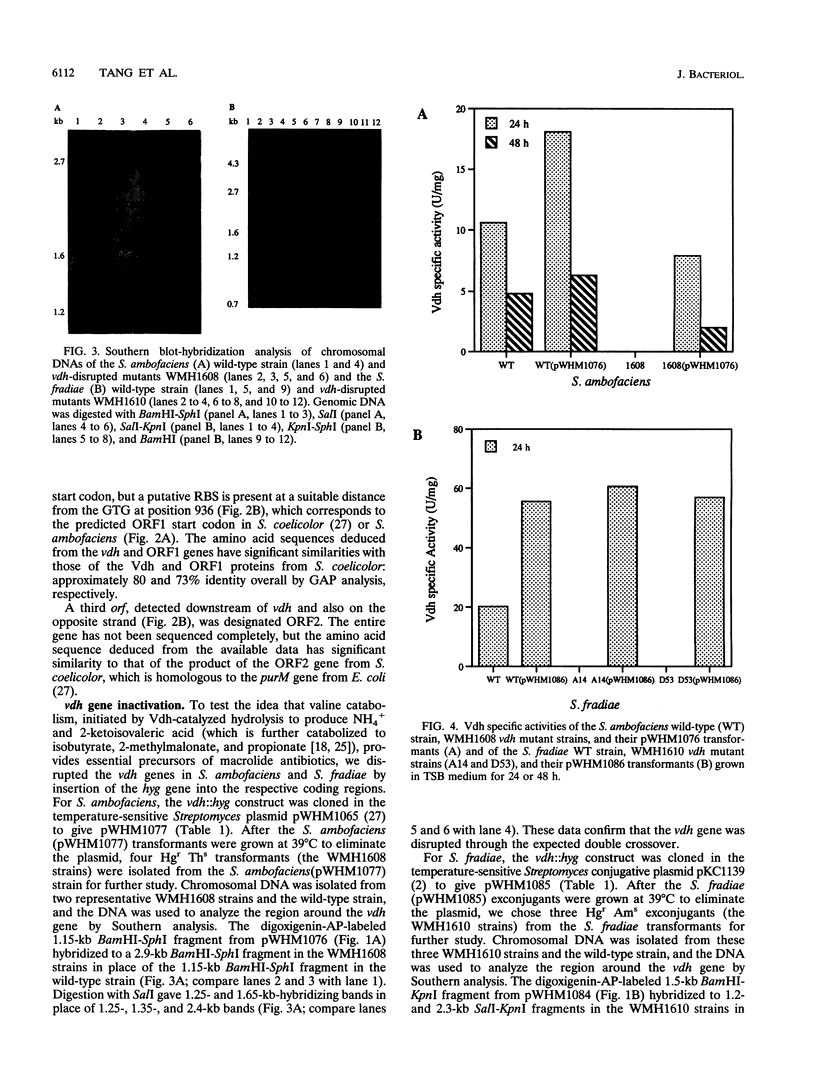
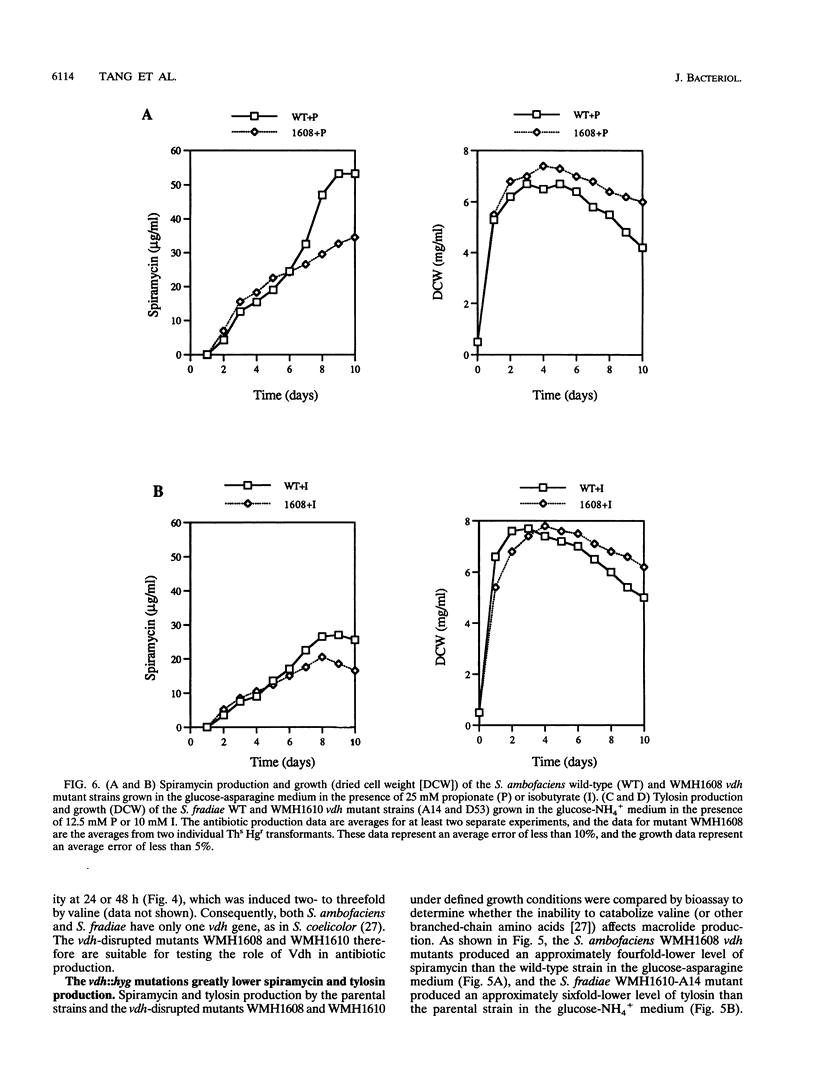
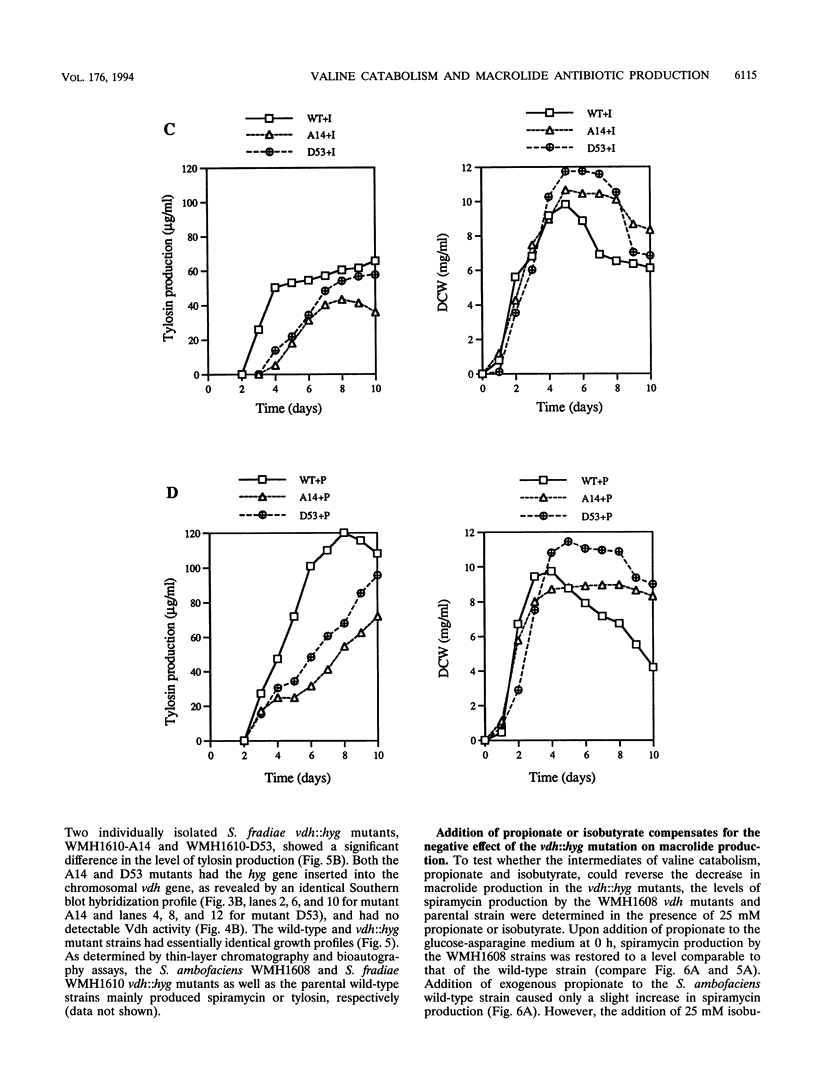
Targeted inactivation of the valine (branched-chain amino acid) dehydrogenase gene (vdh) was used to study the role of valine catabolism in the production of tylosin in Streptomyces fradiae and spiramycin in Streptomyces ambofaciens. The deduced products of the vdh genes, cloned and sequenced from S. fradiae C373.1 and S. ambofaciens ATCC 15154, are approximately 80% identical over all 363 amino acids and 96% identical over a span of the first N-terminal 107 amino acids, respectively, to the deduced product of the Streptomyces coelicolor vdh gene. The organization of the regions flanking the vdh genes is the same in all three species. Inactivation of the genomic copy of the vdh gene in S. fradiae and S. ambofaciens by insertion of a hygromycin resistance (hyg) gene caused loss of the valine dehydrogenase (Vdh) activity, and thus only one enzyme is responsible for the Vdh activity in these organisms. Analysis of the culture broth by bioassay revealed that the vdh::hyg mutants produce an approximately sixfold-lower level of tylosin and an approximately fourfold-lower level of spiramycin than the wild-type S. fradiae and S. ambofaciens strains, while maintaining essentially identical growth in a defined minimal medium with either 25 mM ammonium ion or 0.05% asparagine as the nitrogen source. The addition of the valine catabolite, propionate or isobutyrate, and introduction of the wild-type vdh gene back to each vdh::hyg mutant reversed the negative effect of the vdh::hyg mutation on spiramycin and tylosin production. These data show that the catabolism of valine is a major source of fatty acid precursors for macrolide biosynthesis under defined growth conditions and imply that amino acid catabolism is a vital source of certain antibiotic precursors in actinomycetes.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Bierman M., Logan R., O'Brien K., Seno E. T., Rao R. N., Schoner B. E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992 Jul 1;116(1):43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Decker H., Summers R. G., Hutchinson C. R. Overproduction of the acyl carrier protein component of a type II polyketide synthase stimulates production of tetracenomycin biosynthetic intermediates in Streptomyces glaucescens. J Antibiot (Tokyo) 1994 Jan;47(1):54–63. doi: 10.7164/antibiotics.47.54. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotzlaf J. E., Metzger L. S., Foglesong M. A. Incorporation of amino acid-derived carbon into tylactone by Streptomyces fradiae GS14. Antimicrob Agents Chemother. 1984 Feb;25(2):216–220. doi: 10.1128/aac.25.2.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D. R., Solenberg P. J., Baltz R. H. Tn5099, a xylE promoter probe transposon for Streptomyces spp. J Bacteriol. 1991 Sep;173(17):5573–5577. doi: 10.1128/jb.173.17.5573-5577.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y. J., Kolattukudy P. E. Inhibition of erythromycin synthesis by disruption of malonyl-coenzyme A decarboxylase gene eryM in Saccharopolyspora erythraea. J Bacteriol. 1994 Feb;176(3):714–724. doi: 10.1128/jb.176.3.714-724.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrihi A., Lamsaif D., Lefebvre G., Germain P. Effect of ammonium ions on spiramycin biosynthesis in Streptomyces ambofaciens. Appl Microbiol Biotechnol. 1992 Jun;37(3):382–387. doi: 10.1007/BF00210997. [DOI] [PubMed] [Google Scholar]
- Malmberg L. H., Hu W. S., Sherman D. H. Precursor flux control through targeted chromosomal insertion of the lysine epsilon-aminotransferase (lat) gene in cephamycin C biosynthesis. J Bacteriol. 1993 Nov;175(21):6916–6924. doi: 10.1128/jb.175.21.6916-6924.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin J. F., Demain A. L. Control of antibiotic biosynthesis. Microbiol Rev. 1980 Jun;44(2):230–251. doi: 10.1128/mr.44.2.230-251.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey L. K., Sokatch J. R., Conrad R. S. Branched-chain amino acid catabolism in bacteria. Bacteriol Rev. 1976 Mar;40(1):42–54. doi: 10.1128/br.40.1.42-54.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez C., Braña A. F., Manzanal M. B., Hardisson C. Role of substrate mycelium in colony development in Streptomyces. Can J Microbiol. 1985 May;31(5):446–450. doi: 10.1139/m85-083. [DOI] [PubMed] [Google Scholar]
- Navarrete R. M., Vara J. A., Hutchinson C. R. Purification of an inducible L-valine dehydrogenase of Streptomyces coelicolor A3(2). J Gen Microbiol. 1990 Feb;136(2):273–281. doi: 10.1099/00221287-136-2-273. [DOI] [PubMed] [Google Scholar]
- Omura S., Taki A., Matsuda K., Tanaka Y. Ammonium ions suppress the amino acid metabolism involved in the biosynthesis of protylonolide in a mutant of Streptomyces fradiae. J Antibiot (Tokyo) 1984 Nov;37(11):1362–1369. doi: 10.7164/antibiotics.37.1362. [DOI] [PubMed] [Google Scholar]
- Omura S., Tanaka Y., Mamada H., Masuma R. Effect of ammonium ion, inorganic phosphate and amino acids on the biosynthesis of protylonolide, a precursor of tylosin aglycone. J Antibiot (Tokyo) 1984 May;37(5):494–502. doi: 10.7164/antibiotics.37.494. [DOI] [PubMed] [Google Scholar]
- Omura S., Tsuzuki K., Tanaka Y., Sakakibara H., Aizawa M., Lukacs G. Valine as a precursor of n-butyrate unit in the biosynthesis of macrolide aglycone. J Antibiot (Tokyo) 1983 May;36(5):614–616. doi: 10.7164/antibiotics.36.614. [DOI] [PubMed] [Google Scholar]
- Priestley N. D., Robinson J. A. Purification and catalytic properties of L-valine dehydrogenase from Streptomyces cinnamonensis. Biochem J. 1989 Aug 1;261(3):853–861. doi: 10.1042/bj2610853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M. M., Yue S., Hutchinson C. R. Biosynthesis of lasalocid A. Metabolic interrelationships of carboxylic acid precursors and polyether antibiotics. J Antibiot (Tokyo) 1986 Aug;39(8):1135–1143. doi: 10.7164/antibiotics.39.1135. [DOI] [PubMed] [Google Scholar]
- Tang L., Hutchinson C. R. Sequence, transcriptional, and functional analyses of the valine (branched-chain amino acid) dehydrogenase gene of Streptomyces coelicolor. J Bacteriol. 1993 Jul;175(13):4176–4185. doi: 10.1128/jb.175.13.4176-4185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas D. I., Cove J. H., Baumberg S., Jones C. A., Rudd B. A. Plasmid effects on secondary metabolite production by a streptomycete synthesizing an anthelmintic macrolide. J Gen Microbiol. 1991 Oct;137(10):2331–2337. doi: 10.1099/00221287-137-10-2331. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Vancura A., Vancurová I., Volc J., Fussey S. P., Flieger M., Neuzil J., Marsálek J., Behal V. Valine dehydrogenase from Streptomyces fradiae: purification and properties. J Gen Microbiol. 1988 Dec;134(12):3213–3219. doi: 10.1099/00221287-134-12-3213. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



