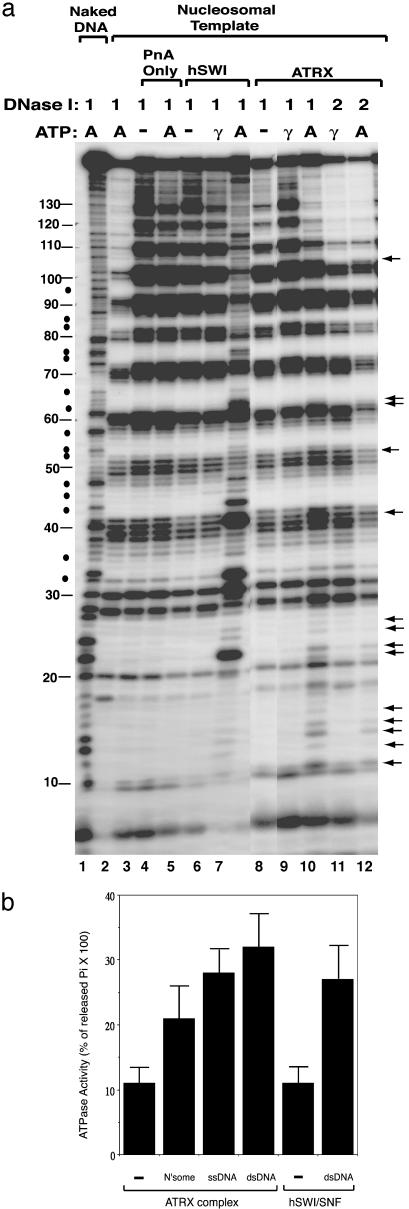
Fig. 3.
The ATRX complex alters the DNase I digestion pattern of a nucleosome in the presence of ATP. (a) Autoradiograph showing the results of the mononucleosome disruption assay. Complexes isolated by protein A alone (PnA) or Abs against ATRX and human SWI/SNF (hSWI) are indicated at the top. The templates used and the presence of ATP (A) or ATP-γ-S (γ) are indicated. The amounts of DNase used, 1 and 2, represent 0.2 and 0.4 units, respectively. The solid arrows mark the nucleotides whose digestion was enhanced by the ATRX–Daxx complex in the presence of ATP. The solid dots denote the nucleotides between the 10-bp ladders whose digestion was stimulated by human SWI/SNF but not ATRX–Daxx. The solid lines mark the positions of the nucleotides in 5S DNA. The 10-bp ladders can be observed between nucleotides 10 and 130 of 5S DNA. (b) Graphic presentation showing that the ATRX complex has DNA- or nucleosome-stimulated ATPase activity (shown in percentage of released inorganic phosphate from total ATP multiplied by 100). The ATPase activity of mock immunoprecipitation (using preimmune serum) is indistinguishable from that of the background (using protein A beads), which was subtracted during calculation. The presence of nucleosomes (N′some), single-stranded DNA (ssDNA), and double-stranded DNA (dsDNA) is indicated.