Figure 5. The loss of pgcA uncouples chromosome segregation from division but does not disrupt nucleoid segregation or DNA replication.
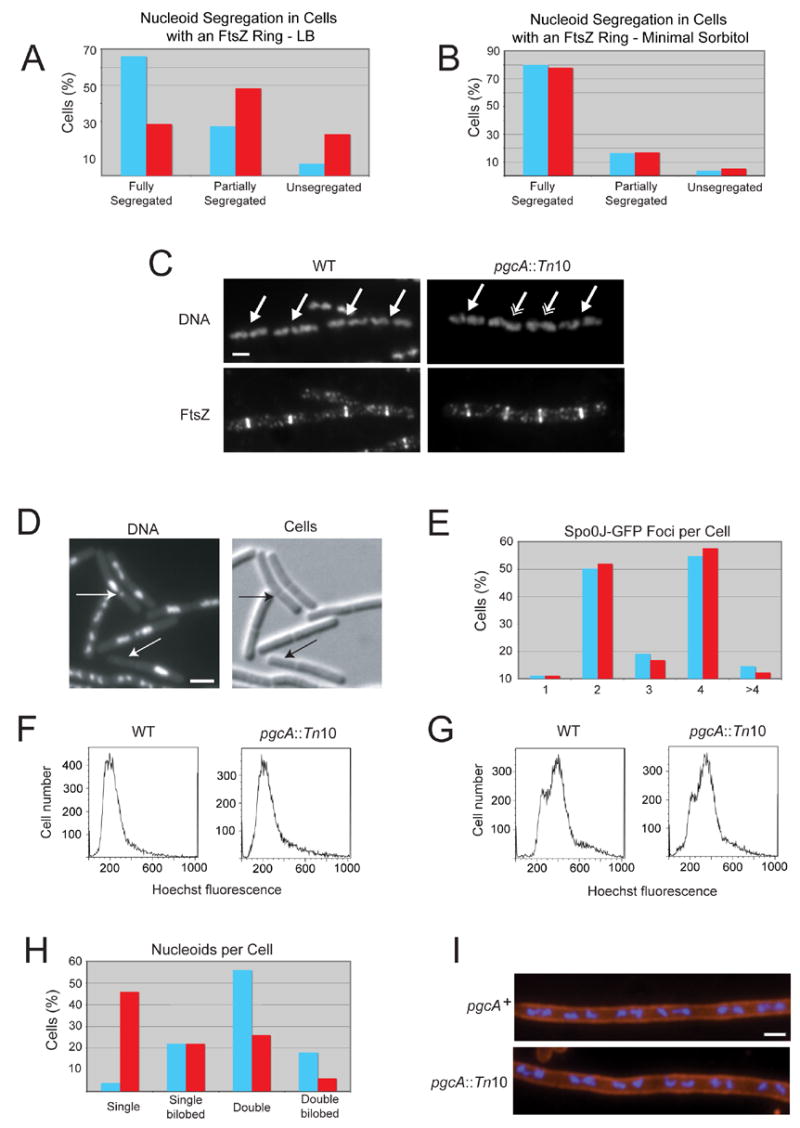
(A and B) The loss of pgcA promotes the formation of FtsZ rings over unsegregated nucleoids in LB (n=126) but not minimal sorbitol (n=59) relative to wild type cells (n=106 for LB, n=55 for minimal sorbitol, p < 0.001). (C) DNA (top) and FtsZ (bottom) in wild type (left) and pgcA::Tn10 (right) cells. Single arrows indicate FtsZ rings over segregated nucleoids. Double arrows indicate FtsZ rings over either unsegregated or partially segregated nucleoids. Bar = 2 μm. (D) Micrographs of pgcA::Tn10 spoIIIE::spc cells displaying chromosome bisection and anucleate phenotypes. Arrows indicate either bisected chromosomes or anucleate cells. Bar = 2 μm. (E) Histogram showing the number of foci of the origin binding protein Spo0J-GFP in wild type (blue, n=87) and pgcA::Tn10 (red, n=88) cells. (F and G) Flow cytometry of wild type and pgcA::Tn10 cells grown in the absence (F) or presence (G) of rifampicin to block replication initiation and cell division indicate that DNA replication is proceeding normally in the mutant strain. (H) Histogram indicating the number and conformation of nucleoids per cell for wild type (blue, n=50) and pgcA::Tn10 (red, n=50) cells. The reduced number of nucleoids in pgcA mutants suggests a delay in nucleoid segregation due to their short stature. (I) pgcA+ and pgcA::Tn10 cells depleted for FtsZ were stained for cell wall (red) and DNA (blue). Length per nucleoid was identical for both strains (~2.5 μm per nucleoid) indicating that nucleoid segregation is normal in the absence of septation in pgcA mutant cells.
