Fig. 2.
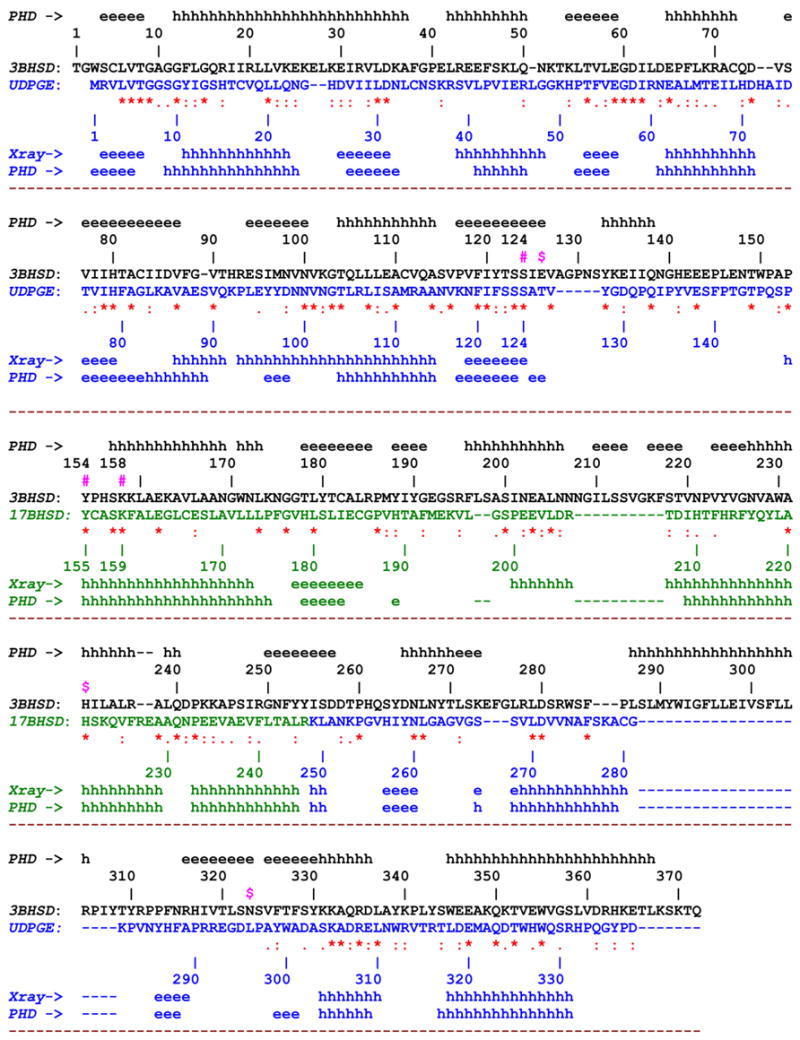
The 3D amino acid sequence alignment of 3β-HSD_1 with corresponding constituent parts of UDPGE and 17β-HSD_1. The experimental (X-ray) and predicted (PHD method [16]) secondary structure elements are indicated along the sequence The sequences and individual numbering of 3β-HSD_1, UDPGE and 17β-HSD_1 are shown in black, blue and green respectively. The yellow color marks the region, deletion of which does not cause the loss of enzyme activity.
Other indicative symbols:
#(in magenta) - oxidoreductase catalytic residues, Ser124, Tyr154, Lys158
^ (in blue) - residues lining the NAD binding shell
x (in magenta) – residues lining the substrate binding shell
$ (in magenta) – proposed 3D structure based isomerase catalytic (Glu126) and substrate recognition/binding (His232, Asn323) residues in the active site.
