Abstract
Background
Laboratory methods for HER2 assessment currently include immunohistochemical (IHC) methods (measuring protein overexpression) and fluorescence in situ hybridisation (FISH) (measuring gene amplification). The measure of HER2 protein by IHC is usually assessed by the mouse monoclonal antibody CB11, and polyclonal antibodies (Herceptest) directed against the internal portion of the receptor. Recently, chromogenic in situ hybridisation (CISH), in which HER2 is detected by a peroxidase reaction and the gene amplification can be determined by regular bright‐field microscopy, has emerged as an alternative to FISH.
Aims
To evaluate the status of HER2 in tissue microarrays (TMAs) of invasive breast cancer using the novel rabbit monoclonal antibody SP3 directed against the external portion of HER2, and correlate the results with CB11 and CISH.
Methods
IHC was performed with two antibodies (CB11 and SP3) and CISH for HER2 in 10 TMA blocks with 190 formalin‐fixed paraffin‐embedded cases of invasive breast carcinomas.
Results
The correlation between SP3 and CB11 was significant (p<0.001) with an agreement rate of 86.9%. When the staining pattern of the two antibodies was compared, the majority of SP3 immunostainings were assessed more easily, with a strong complete membrane staining pattern without non‐specific cytoplasmic staining. There was a good correlation between SP3 and CISH (p<0.001). 23/24 SP3 3+ cases showed gene amplification, 97.3% of the cases without gene amplification were SP3 negative, and 6/7 SP3 2+ were amplified.
Conclusion
The high level of agreement between SP3, a monoclonal antibody that recognises the extracellular domain of the HER2 receptor, and CB11 and CISH, shows that this novel antibody is a reliable candidate to evaluate the expression of HER2 in breast cancer.
Keywords: HER2, CISH, SP3, TMA, breast cancer
HER2/neu is a proto‐oncogene mapped to chromosome 17 (17q21) that encodes a transmembrane growth factor receptor with tyrosine kinase activity.1,2 This receptor is overexpressed in 15–30% of invasive breast carcinomas3,4,5,6 and is associated with poor prognosis and resistance to hormonal therapy.5 Overexpression of the HER2 protein and/or amplification of the gene is an eligibility requirement for trastuzumab therapy, a target‐specific therapy that acts by blocking the extracellular domain of the receptor.7
Currently laboratory methods for HER2 assessment include immunohistochemistry (IHC) (measuring protein overexpression) and fluorescence in situ hybridisation (FISH) (measuring gene amplification). Because IHC assessment of HER2 is practical, inexpensive and easily automated, it is the most commonly applied method in pathology laboratories to assess HER2 protein overexpression.
Despite the advantages of IHC, extremely variable results are found in the literature.7,8 Therefore, the standardisation of IHC methodology and the interpretation of results have been strongly recommended by different groups.7,8 Both sensibility and specificity of the antibodies chosen to evaluate HER2 expression are of paramount importance to overcome this variability.
Several commercially available antibodies recognise distinct intracellular or extracellular epitopes of the HER2 molecule, for example, antibodies directed against the intracytoplasmic domain of the protein, specifically the polyclonal antibody (rabbit anti‐human HER2 protein) included in the HercepTest, and the monoclonal antibody CB11 (Novocastra Laboratories Ltd, Newcastle upon Tyne, UK). The monoclonal antibody TAB250 (Novocastra Laboratories Ltd) recognises the extracellular domain of HER2.9,10
SP3 (Labvision Corporation–NeoMarkers, Fremont, California, USA) is a novel rabbit monoclonal antibody directed to the extracellular domain of the HER2 receptor. Since therapy with trastuzumab targeted the extramembrane epitope of HER2, antibodies detecting this portion of the receptor could produce results with higher clinical relevance related to therapy response. Another advantage is that rabbit monoclonal antibodies are a category of immunoreagents that combine the best properties of both mouse monoclonal antibodies and rabbit antisera, having a good sensibility and specificity of staining.11,12,13
Despite this diversity of antibodies, UK pathologists recommend the use of the FDA‐approved antibodies and scoring system to accomplish the standardisation of IHC methodology and interpretation of the results to evaluate HER2.14 Nowadays, the graduation system of IHC for HER2 is based on intensity and extension of the membrane staining,14,15 being HercepTest and CB11, the only FDA‐approved antibodies. The eligible parameters for treatment with Herceptin are the IHC 3+ score and/or gene amplification measurable by in situ hybridisation.14
FISH is the universally accepted gold standard method for confirming IHC 2+ cases and ambiguous results, but it is expensive and requires technical expertise. Nevertheless, this technique needs specific laboratory equipment and fluorescent signals quickly fade, which means that FISH slides cannot be stored permanently. Recently, chromogenic in situ hybridisation (CISH), which enables detection of HER2 gene copies by conventional peroxidase reaction using bright field microscopy evaluation, has been proposed as an alternative to FISH.16,17,18,19,20,21 Several comparative studies have shown an overall good agreement between CISH and FISH (84–100%),16,17,21,22,23,24,25,26,27 showing that HER2 status can be reliably assessed by CISH. Gon et al, studying 80 cases of invasive breast carcinomas, showed near‐perfect agreement between FISH and CISH (91%) when evaluated by three pathologists.21 An excellent concordance (94.8%) between CISH and FISH was shown by Saez et al: sensitivity of CISH was 97.5% and specificity 94%, considering FISH as gold standard.18 CISH and FISH correlated well in a series of 157 breast cancers (κ 0.81) studied by Tanner et al.16 The few discrepancies were mostly because of low‐level amplifications by FISH that were negative by CISH. The aim of this study was to evaluate the status of HER2 in a series of 190 formalin‐fixed paraffin‐embedded invasive breast carcinomas, arrayed in 10 tissue microarray (TMA) blocks, using the novel rabbit monoclonal antibody SP3, in order to correlate its sensibility and specificity with the mouse monoclonal antibody CB11 and the gene copy number status performed by CISH.
Materials and methods
Specimen selection and TMA construction
A total of 119 cases of invasive breast carcinoma sent for evaluation of HER2 gene status were selected from the files of the Pathology Unit of IPATIMUP (Institute of Molecular Pathology and Immunology of Porto University). Ten TMAs were constructed from the archived formalin‐fixed, paraffin‐embedded resection blocks using the “TMA Builder” (G Szekeres, #WO 2004/000992, Histopathology Ltd, Hungary). One representative area from each breast carcinoma was defined from corresponding H&E‐stained sections and marked. The 2.0 mm diameter cores were sampled from these areas in the donor block and inserted into an empty recipient in the receptor block in an ordered manner. Consecutive 3–4 μm sections were then cut on to adhesive‐coated slides, baked overnight at 37°C, and subsequently processed by CISH and immunohistochemistry.
Immunohistochemistry
IHC for HER2 protein was done in 3 μm TMA tissue sections. Two monoclonal antibodies were used in this study: NCL‐CB11, recognising the intracellular portion (mouse monoclonal antibody, Novocastra Laboratories Ltd), and SP3, recognising the extracellular portion (rabbit monoclonal antibody, Labvision Corporation–NeoMarkers). Tissue sections were deparaffinised followed by antigen‐retrieval in citrate buffer (0.01 M pH 6.0) at high temperature (water bath, 30 min at 98°C). After blocking for non‐specific binding, primary antibody was added on the sections in a pre‐standardised concentration (CB11 1/60; SP3 1/80) and incubated (CB11 and SP3, 30 min at room temperature). A standard avidin–biotin–peroxidase complex technique was used for visualisation, with diaminobenzidine as the chromogen (UltraVision Detection System Anti‐Polyvalent, HRP/DAB (Ready‐To‐Use), LabVision Corporation). TMA tissue sections were lightly counterstained with haematoxylin and cover‐slipped.
For CB11 and SP3, the Herceptest scoring system was applied: negative, no membrane staining or <10% of cells stained; 1+, incomplete membrane staining in >10% of cells; 2+, >10% of cells with weak to moderate complete membrane staining; and 3+, strong and complete membrane staining in >10% of cells. Appropriate HER2 overexpressed breast carcinomas controls were included in each run; each TMA section was analysed by two of the authors (FM and FCS) independently.
CISH
Consecutive 4 μm sections were cut from the 10‐TMA block on to adhesive‐coated slides and baked overnight in a 37°C oven. Briefly, the sections were deparaffinised, followed by heat pretreatment in Pre‐treatment Buffer (Spot‐Light Tissue Pretreatment Kit, Zymed, California, USA) and pepsin digestion. Ready‐to‐use digoxigenin‐labelled HER2 probe (Spot‐Light HER2 probe, Zymed) was applied on to slides before denaturation and hybridisation. After the stringent wash, the HER2 probe was detected with sequential incubation with fluorescein isothiocyanate (FITC) conjugated anti‐digoxigenin antibody, HRP‐conjugate anti‐FITC antibody, and DAB according to the direction on the manufacture's specification data sheet (CISH Detection Kit, Zymed).
To validate the reliability of the HER2 gene amplification and exclusion of the possibility of aneuploidy, we performed the chromosome 17 probe using the Spot‐Light Zymed probe in every TMA tissue section, in accordance with the manufacturer's protocol. The biotin‐labelled chromosome 17 centromere probe (Zymed) was detected with sequential incubation with HRP‐conjugated streptavidin and DAB. TMA tissue sections were lightly counterstained with haematoxylin and cover‐slipped. Chromosome 17 CISHM was interpreted as disomy or aneusomy and numbers were compared to HER2 to define true amplification, as previously recommended.26 Appropriate gene amplified breast carcinoma controls were included in each run and the TMA section was analysed by two of the authors (FM and SR) independently. Signals were evaluated at 400× and 630× and the tumour was considered to be amplified for HER2 when more than 50% of the neoplastic cells exhibited more than five signals per nucleus or large gene signal clusters.27
Results
The correlation between the two antibodies, SP3 (directed to the extracellular portion of the HER2 receptor) and CB11 (directed to the intracellular portion), was statistically significant (p<0.001) with an agreement rate of 86.9% (table 1).
Table 1 Correlation between the antibodies CB11 and SP3.
| CB11 | SP3 | Total | |||
|---|---|---|---|---|---|
| 0 | + | ++ | +++ | ||
| 0 | 138 (72.6%) | 1 (0.5%) | 0 (0.0%) | 1 (0.5%) | 140 (73.7%) |
| + | 7 (3.7%) | 3 (1.6%) | 1 (0.5%) | 2 (1.1%) | 13 (6.8%) |
| ++ | 5 (2.6%) | 2 (1.1%) | 2 (1.1%) | 1 (0.5%) | 10 (5.3%) |
| +++ | 1 (0.5%) | 0 (0.0%) | 4 (2.1%) | 22 (11.6%) | 27 (14.2%) |
| Total | 151 (79.5%) | 6 (3.2%) | 7 (3.7%) | 26 (13.7%) | 190 (100%) |
χ2 : p<0.001.
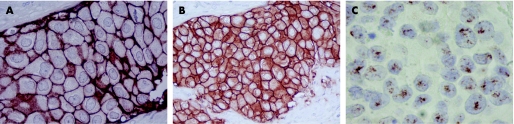
Our results show a similar number of cases scored as 3+ with SP3 and CB11 (SP3 3+ = 13.7%; CB11 3+ = 14.2%). On the other hand, SP3 2+ was observed in 3.7% of cases, while CB11 2+ was observed in 5.3%. When cases scored as negative or 1+ are grouped and cases scored as 2+ are excluded, the correlation remains, with four discordant cases: one case CB11 3+ was negative with SP3, and was not amplified; three cases were CB11 negative and considered SP3 3+, in which two were amplified and one was not amplified. When we compare the staining pattern of the two antibodies, we observe that the majority of SP3 immunostainings (fig 1A) were assessed more easily, with a strong complete membrane staining pattern without non‐specific cytoplasmic staining. On the other hand, CB11 immunostaining (fig 1B) frequently showed non‐specific cytoplasmic staining, which leads to a greater interobserver variability. The interobserver analysis show a discordance rate of 9.5% in SP3 immunostaining and 14.7% with CB11. However, clinically relevant discordances were observed in five cases for both antibodies.
Figure 1 HER2 overexpression and gene amplification in invasive breast cancer arrayed in TMA. (A) SP3 3+ immunostaining, 400×; (B) CB11 3+ immunostaining, 400×; (C) CISH showing HER2 amplification, 630×.
Since in the literature, high correlation rates were shown between gene amplification assessment by FISH and CISH,15,16,17,21,22,23,24,25,26,27 we consider CISH as a gold standard and compare the results of gene amplification with this technique and ICH results with CB11 and SP3. Among the 190 cases tissue‐arrayed, 29 were excluded from the analysis because of inconclusive results for CISH for several reasons: detachment of cores, few invasive neoplastic cells for analysis, no staining, etc. All the cases analysed were diploid for chromosome 17. Table 2 shows that the correlation between SP3 and CISH was highly significant (p<0.001). In 98.2% of the cases that were negative or 1+ for SP3, CISH did not show gene amplification. On the other hand, among 24 cases showing 3+ immunoreactivity for SP3, 23 had gene amplification for CISH. One interesting result is that of seven SP3 2+ cases, six showed gene amplification. This result contrasts with those observed for CB11, where only 6/10 2+ cases showed amplification (table 3).
Table 2 Correlation between SP3 and chromogenic in situ hybridisation (CISH).
| CISH | SP3 | Total | |||
|---|---|---|---|---|---|
| 0 | + | ++ | +++ | ||
| Negative | 108 (97.3%) | 1 (0.9%) | 1 (0.9%) | 1 (0.9%) | 111 (68.9%) |
| Positive | 16 (32.0%) | 5 (10.0%) | 6 (12.0%) | 23 (46.0%) | 50 (31.1%) |
| Total | 124 (77.0%) | 6 (3.7%) | 7 (4.4%) | 24 (14.9%) | 161 (100%) |
χ2 : p<0.001.
Table 3 Correlation between CB11 and chromogenic in situ hybridisation (CISH).
| CISH | CB11 | Total | |||
|---|---|---|---|---|---|
| 0 | + | ++ | +++ | ||
| Negative | 100 (90.1%) | 5 (4.5%) | 4 (3.6%) | 2 (1.8%) | 111 (68.9%) |
| Positive | 14 (28.0%) | 7 (14.0%) | 6 (12.0%) | 23 (46.0%) | 50 (31.1%) |
| Total | 114 (70.8%) | 12 (7.5%) | 10 (6.2%) | 25 (15.5%) | 161 (100%) |
χ2 : p <0.001.
In the overall assessment, CB11 results were also correlated significantly when compared with CISH (p<0.001). In 94.6% of the negative or 1+ cases, CISH was also negative. Among the 25 cases showing CB11 3+ immunoreactivity, 23 had gene amplification.
Table 4 shows that both antibodies revealed high specificity (98%) but relatively low sensitivity, when compared to CISH evaluation: 52% for SP3 and 52% for CB11.
Table 4 Sensibility, specificity, positive predictive value (PPV) and negative predictive value (NPV) of immunohistochemistry using SP3 and CB11 antibodies, having chromogenic in situ hybridisation (CISH) as gold standard.
| Sensibility | Specificity | PPV | NPV | |
|---|---|---|---|---|
| CB11 | 0.52 | 0.98 | 0.92 | 0.93 |
| SP3 | 0.52 | 0.99 | 0.96 | 0.84 |
Discussion
HER2 status can be detected by several methods, analysing the number of gene copies (Southern blotting, PCR or FISH) or protein expression (western blotting, enzyme immunoassay or IHC). In pathology laboratories, the two most commonly used methods are FISH and IHC since they are able to assess HER2 status in formalin‐fixed paraffin‐embedded tissues.
IHC is the most commonly used tool in pathology laboratories for diagnostic purposes. However, the protein evaluation through this procedure is overwhelmed by technical artefacts, sensitivity discrepancies between different antibodies, and interobserver variability between pathologists' interpretations.28 Studies reveal that the interobserver agreement is poor in cases of IHC staining intensity of 1+ and 2+, and the predictive value is unsatisfactory for clinical use; therefore they recommend additional testing by FISH.29
FISH is considered the gold standard method for HER2 evaluation.7 However, this procedure has its disadvantages: it is an expensive and sophisticated method; it needs a fluorescence microscope; and the signal is transitory. CISH is a new detection method of in situ hybridisation, similar to the detection system used in the IHC technique. Since Tanner et al16 described this method, several authors have reported the advantages of CISH over FISH.15,16,17,21,22,23,24,25,26,27,30,31,32 CISH requires an ordinary light microscope; the cost effectiveness of the method is better; the signal intensity is permanent; and the pathologists are familiar with IHC staining, being able to better correlate findings with the underlying tumour morphology.30,32
Despite the advantages of in situ hybridisation methodologies, IHC is still the technique widely used for assessment of HER2 status in pathology laboratories. Of clinical relevance is how to choose the best antibody to evaluate HER2 status.7,33 In the present work, we used a series of 190 formalin‐fixed paraffin‐embedded invasive breast carcinomas, arrayed in 10‐TMA blocks, to measure the sensitivity and specificity of the novel rabbit monoclonal antibody SP3 and the presence of the external domain of HER2 receptor.
Our results show that the pattern of staining of SP3 3+ score was specific, taking into account the parameters of evaluation by the Herceptest scoring system. In fact, the SP3 immunostaining enhances the membrane staining, allowing an unequivocal evaluation of HER2 surface protein, in contrast with the CB11 staining pattern, which shows unspecific cytoplasmic staining.
SP3 staining had a high specificity (99.1%) but a moderate sensibility (52.3%) to assess HER2, having CISH as a gold standard. These moderate results in sensibility are due to the high level of cases SP3 negative/1+ that were amplified by CISH. The possible cleavage of HER2 with loss of the extracellular domain may explain the high number of false negative cases observed with SP3. The shedding of SP3 binding site might be responsible for the negative/1+ IHC classification, despite the presence of gene amplification. One can hypothesise that the extracellular domain‐directed antibodies constitute an excellent tool to predict the availability of the trastuzumab target epitope. It is essential to predict trastuzumab activity in order to select patients whose tumour cells maintain the target epitope. Recently, Bussolati et al34 described a technique, based on the biotinylation of trastuzumab (biotHER), which allows the use of the anti‐HER2 humanised antibody for IHC. Thus, BiotHER can be used as a primary antibody when evaluating the availability of trastuzumab‐specific binding site in breast cancer tissue sections. In addition, BiotHER positivity was a strong predictor of clinical outcome in patients with advanced breast cancer treated with trastuzumab and chemotherapy.34 However, further studies are necessary to confirm these findings. Meanwhile, since SP3 2+ and 3+ cases were greatly associated with gene amplification and SP3 does recognise the ECD of HER2, it should be also investigated in the future as a better predictor of trastuzumab response than the other conventional antibodies used routinely.
TMA is a new tool in histopathology and its reliability to evaluate HER2 protein expression and gene amplification has been discussed.35 This approach allows the evaluation of a greater number of cases in a less costly and tissue‐consuming manner. However, is important to ensure a similar fixation/processing of the samples to achieve better results in IHC and in situ hybridisation methods. In our series we found a high number of CISH inconclusive cases (15.3%). The causes of testing failure and false negative results offered by previous CISH studies include an absence of tumour on section, an inability to score owing to high background, a low signal intensity, an absence of signal from the internal control, and the use of inappropriate fixative.17,24,27,30,36 It was further noted that heat pretreatment and digestion with pepsin are the most critical procedures for optimised CISH performance, and that successful rates of CISH were low when old blocks were used.27 In our study we used paraffin blocks from different institutions to build a TMA block which does not allow the adjustment of digestion times to each sample, since they are located in the same slide. These facts can also be applicable to IHC results. Therefore, we may conclude that for more precise results, TMA blocks must be designed with uniformly fixed and processed samples.
Take‐home messages
The high level of agreement between SP3, a rabbit monoclonal antibody that recognises the extracellular domain of the HER2 receptor, and CB11 and chromogenic in situ hybridisation (CISH), shows that this novel antibody is a reliable candidate to evaluate the expression of HER2 in breast cancer cases.
SP3 staining had a high specificity (99.1%) but a moderate sensibility (52.3%) to assess HER2, having CISH as a gold standard.
The assessment of SP3 and other antibodies that recognise the extracellular domain of HER2, could constitute an excellent tool to predict the availability of the trastuzumab target epitope.
In conclusion, the high level of agreement between SP3, a monoclonal antibody that recognises the extracellular domain of the HER2 receptor, and CB11 and CISH, shows that this novel antibody is a reliable candidate to evaluate the expression of HER2 in breast cancer cases. However, further studies are required to confirm if SP3 could be a better predictor of patient response to trastuzumab therapy than the antibodies that recognise the internal domain of HER2.
Abbreviations
CISH - chromogenic in situ hybridisation
FISH - fluorescence in situ hybridisation
IHC - immunohistochemistry
TMA - tissue microarray
Footnotes
Funding: This work was partially supported by INOPAT (Portugal). SC is recipient of a fellowship grant from FCT (SFRH/BD/21551/2005).
Competing interests: None declared.
References
- 1.Popescu N C, King C R, Kraus M H. Localization of the human erbB‐2 gene on normal and rearranged chromosomes 17 to bands q12–21.32. Genomics 19894362–366. [DOI] [PubMed] [Google Scholar]
- 2.Coussens L, Yang‐Feng T L, Liao Y C.et al Tyrosine kinase receptor with extensive homology to EGF receptor shares chromosomal location with neu oncogene. Science 19852301132–1139. [DOI] [PubMed] [Google Scholar]
- 3.Olayioye M A, Neve R M, Lane H A.et al The ErbB signaling network: receptor heterodimerization in development and cancer. Embo J 2000193159–3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schechter A L, Stern D F, Vaidyanathan L.et al The neu oncogene: an erb‐B‐related gene encoding a 185,000‐Mr tumour antigen. Nature 1984312513–516. [DOI] [PubMed] [Google Scholar]
- 5.Slamon D J, Clark G M, Wong S G.et al Human breast cancer: correlation of relapse and survival with amplification of the HER‐2/neu oncogene. Science 1987235177–182. [DOI] [PubMed] [Google Scholar]
- 6.Hudelist G, Kostler W J, Attems J.et al Her‐2/neu‐triggered intracellular tyrosine kinase activation: in vivo relevance of ligand‐independent activation mechanisms and impact upon the efficacy of trastuzumab‐based treatment. Br J Cancer 200389983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bilous M, Dowsett M, Hanna W.et al Current perspectives on HER2 testing: a review of national testing guidelines. Mod Pathol 200316173–182. [DOI] [PubMed] [Google Scholar]
- 8.Ellis I O, Dowsett M, Bartlett J.et al Recommendations for HER2 testing in the UK. J Clin Pathol 200053890–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Corbett I P, Henry J A, Angus B.et al NCL‐CB11, a new monoclonal antibody recognizing the internal domain of the c‐erbB‐2 oncogene protein effective for use on formalin‐fixed, paraffin‐embedded tissue. J Pathol 199016115–25. [DOI] [PubMed] [Google Scholar]
- 10.Ceccarelli C, Santini D, Gamberini M.et al Immunohistochemical expression of internal and external ErbB‐2 domains in invasive breast cancer. Breast Cancer Res Treat 199958107–114. [DOI] [PubMed] [Google Scholar]
- 11.Verbanac K M, Gross U, Rebellato L M.et al Generation of rabbit anti‐lymphocyte monoclonal antibodies. Transplant Proc 199325837–838. [PubMed] [Google Scholar]
- 12.Kuo M C, Sogn J A, Max E E.et al Rabbit‐mouse hybridomas secreting intact rabbit immunoglobulin. Mol Immunol 198522351–359. [DOI] [PubMed] [Google Scholar]
- 13.Verbanac K M, Gross U M, Rebellato L M.et al Production of stable rabbit‐mouse heterohybridomas: characterization of a rabbit monoclonal antibody recognizing a 180 kDa human lymphocyte membrane antigen. Hybridoma 199312285–295. [DOI] [PubMed] [Google Scholar]
- 14.Birner P, Oberhuber G, Stani J.et al Evaluation of the United States Food and Drug Administration‐approved scoring and test system of HER‐2 protein expression in breast cancer. Clin Cancer Res 200171669–1675. [PubMed] [Google Scholar]
- 15.Lewis F, Jackson P, Lane S.et al Testing for HER2 in breast cancer. Histopathology 200445207–217. [DOI] [PubMed] [Google Scholar]
- 16.Tanner M, Gancberg D, Di Leo A.et al Chromogenic in situ hybridization: a practical alternative for fluorescence in situ hybridization to detect HER‐2/neu oncogene amplification in archival breast cancer samples. Am J Pathol 20001571467–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta D, Middleton L P, Whitaker M J.et al Comparison of fluorescence and chromogenic in situ hybridization for detection of HER‐2/neu oncogene in breast cancer. Am J Clin Pathol 2003119381–387. [DOI] [PubMed] [Google Scholar]
- 18.Saez A, Andreu F J, Segui M A.et al HER‐2 gene amplification by chromogenic in situ hybridisation (CISH) compared with fluorescence in situ hybridisation (FISH) in breast cancer—a study of two hundred cases. Breast 200615519–527. [DOI] [PubMed] [Google Scholar]
- 19.Loring P, Cummins R, O'Grady A.et al HER2 positivity in breast carcinoma: a comparison of chromogenic in situ hybridization with fluorescence in situ hybridization in tissue microarrays, with targeted evaluation of intratumoral heterogeneity by in situ hybridization. Appl Immunohistochem Mol Morphol 200513194–200. [DOI] [PubMed] [Google Scholar]
- 20.Bhargava R, Lal P, Chen B. Chromogenic in situ hybridization for the detection of HER‐2/neu gene amplification in breast cancer with an emphasis on tumors with borderline and low‐level amplification: does it measure up to fluorescence in situ hybridization? Am J Clin Pathol 2005123237–243. [PubMed] [Google Scholar]
- 21.Gong Y, Gilcrease M, Sneige N. Reliability of chromogenic in situ hybridization for detecting HER‐2 gene status in breast cancer: comparison with fluorescence in situ hybridization and assessment of interobserver reproducibility. Mod Pathol 2005181015–1021. [DOI] [PubMed] [Google Scholar]
- 22.Vera‐Roman J M, Rubio‐Martinez L A. Comparative assays for the HER‐2/neu oncogene status in breast cancer. Arch Pathol Lab Med 2004128627–633. [DOI] [PubMed] [Google Scholar]
- 23.Arnould L, Gelly M, Penault‐Llorca F.et al Trastuzumab‐based treatment of HER2‐positive breast cancer: an antibody‐dependent cellular cytotoxicity mechanism? Br J Cancer 200694259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dandachi N, Dietze O, Hauser‐Kronberger C. Chromogenic in situ hybridization: a novel approach to a practical and sensitive method for the detection of HER2 oncogene in archival human breast carcinoma. Lab Invest 2002821007–1014. [DOI] [PubMed] [Google Scholar]
- 25.Park K, Kim J, Lim S.et al Comparing fluorescence in situ hybridization and chromogenic in situ hybridization methods to determine the HER2/neu status in primary breast carcinoma using tissue microarray. Mod Pathol 200316937–943. [DOI] [PubMed] [Google Scholar]
- 26.Isola J, Tanner M, Forsyth A.et al Interlaboratory comparison of HER‐2 oncogene amplification as detected by chromogenic and fluorescence in situ hybridization. Clin Cancer Res 2004104793–4798. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Wu R, Au A.et al Determination of HER2 gene amplification by chromogenic in situ hybridization (CISH) in archival breast carcinoma. Mod Pathol 200215657–665. [DOI] [PubMed] [Google Scholar]
- 28.Press M F, Hung G, Godolphin W.et al Sensitivity of HER‐2/neu antibodies in archival tissue samples: potential source of error in immunohistochemical studies of oncogene expression. Cancer Res 1994542771–2777. [PubMed] [Google Scholar]
- 29.Thomson T A, Hayes M M, Spinelli J J.et al HER‐2/neu in breast cancer: interobserver variability and performance of immunohistochemistry with 4 antibodies compared with fluorescent in situ hybridization. Mod Pathol 2001141079–1086. [DOI] [PubMed] [Google Scholar]
- 30.Kumamoto H, Sasano H, Taniguchi T.et al Chromogenic in situ hybridization analysis of HER‐2/neu status in breast carcinoma: application in screening of patients for trastuzumab (Herceptin) therapy. Pathol Int 200151579–584. [DOI] [PubMed] [Google Scholar]
- 31.Vocaturo A, Novelli F, Benevolo M.et al Chromogenic in situ hybridization to detect HER‐2/neu gene amplification in histological and ThinPrep(R)‐processed breast cancer fine‐needle aspirates: a sensitive and practical method in the trastuzumab era. Oncologist 200611878–886. [DOI] [PubMed] [Google Scholar]
- 32.Madrid M A, Lo R W. Chromogenic in situ hybridization (CISH): a novel alternative in screening archival breast cancer tissue samples for HER‐2/neu status. Breast Cancer Res 20046R593–R600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gouvea A P, Milanezi F, Olson S J.et al Selecting antibodies to detect HER2 overexpression by immunohistochemistry in invasive mammary carcinomas. Appl Immunohistochem Mol Morphol 200614103–108. [DOI] [PubMed] [Google Scholar]
- 34.Bussolati G, Montemurro F, Righi L.et al A modified trastuzumab antibody for the immunohistochemical detection of HER‐2 overexpression in breast cancer. Br J Cancer 2005921261–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sapino A, Coccorullo Z, Cassoni P.et al Which breast carcinomas need HER‐2/neu gene study after immunohistochemical analysis? Results of combined use of antibodies against different c‐erbB2 protein domains. Histopathology 200343354–362. [DOI] [PubMed] [Google Scholar]
- 36.Arnould L, Denoux Y, MacGrogan G.et al Agreement between chromogenic in situ hybridisation (CISH) and FISH in the determination of HER2 status in breast cancer. Br J Cancer 2003881587–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]