Figure 1.
Isolation of ESD4.
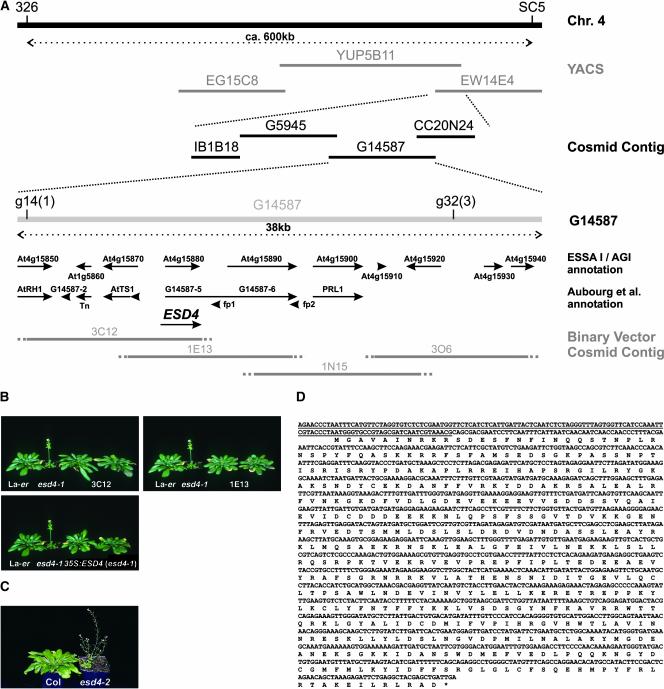
(A) Map-based cloning of ESD4. ESD4 was positioned between cleaved amplified polymorphic sequence (CAPS) markers 326 and SC5 in a segregating F2 population derived from an esd4 (Ler) × Columbia cross. A yeast artificial chromosome (YAC) and a cosmid contig covering this region had been constructed previously (Bevan et al., 1998). Two new CAPS markers, g14 and g32, were generated that flanked esd4 such that one and three crossovers were identified between esd4 and g14 and g32, respectively. Thus, ESD4 was positioned to a region of ∼35 kb, covered by cosmid G14587. This region has been annotated independently by the Arabidopsis Genome Initiative (2000) and Aubourg et al. (1997). Predicted genes from these annotations are shown as arrows. A binary vector cosmid contig covering the region containing ESD4 was constructed from YAC YUP5B11. The exact end point of each cosmid was not known; therefore, these are shown as dotted lines. Four cosmids (3C12, 1E13, 1N15, and 3O6) were transformed into the esd4 mutant via Agrobacterium-mediated root transformation. Cosmids 3C12 and 1E13 complemented the mutation, indicating that ESD4 is located in the overlap of these cosmids. Subsequent analysis indicated that ESD4 corresponds to At4g15880/G14587-5, although the exact sequence is different from the predicted annotations (see below).
(B) Complementation of esd4-1. Photographs of 7-week-old plants grown under short days. Ler and esd4-1 plants are shown to the left of esd4-1 plants transformed with cosmids 3C12, 1E13, or a 35S::ESD4 cDNA construct. Two independent lines are shown for each construct. esd4-1 plants flowered much earlier than wild-type plants. The transformed plants flower at approximately the same time as Ler and show none of the other effects of esd4-1 (Reeves et al., 2002), indicating complementation of the esd4 mutation.
(C) Early-flowering phenotype caused by the esd4-2 allele. Photographs of 7-week-old plants grown under short days. The esd4-2 mutant flowered with 9.8 ± 0.5 rosette leaves and 2.8 ± 0.7 cauline leaves compared with 51.3 ± 4.6 rosette leaves and 9.3 ± 2.8 cauline leaves for the Columbia wild type.
(D) Sequence and annotation of the ESD4 cDNA. A PCR-based approach was used to identify an ESD4 cDNA. The sequence of the ESD4 cDNA is shown, together with the predicted amino acid sequence of the longest open reading frame. This sequence is slightly longer than the Arabidopsis Genome Initiative At4g15880 prediction. The region of the cDNA deleted in the esd4-1 mutant is underlined. The deletion in esd4-1 is 762 bp long and occurs between and including bases 180,698 and 181,459 of ESD4. The sequence likely represents the full-length ESD4 ORF, because the ORF ends in a translational stop, and 27 bp upstream of the predicted AUG initiation codon there is an in-frame UAG translational stop.