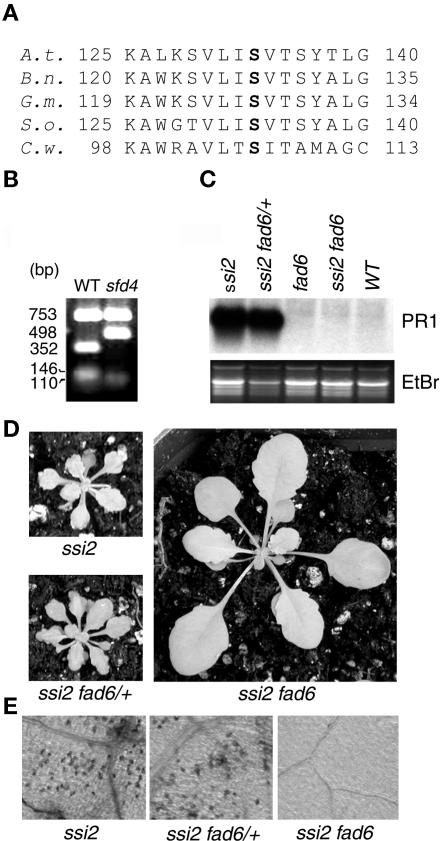
Figure 6.
sfd4 Contains a Mutation in the FAD6 Gene.
(A) Alignment of amino acid sequences in a conserved region of plastidic ω6 desaturases from Arabidopsis thaliana (A.t.), rape (Brassica napus; B.n.), soybean (Glycine max; G.m.), spinach (Spinacia oleracea; S.o.), and the green alga Chlamydomonas sp W80 (C.w.). The Ser-133 that is mutated to Phe-133 in sfd4 is shown in boldface. Numbers at left and right indicate amino acid positions.
(B) EcoRV restriction polymorphism generated by a C→T mutation at the FAD6 locus in the sfd4 ssi2 npr1 plant. PCR-amplified products from the wild type (WT) and sfd4 ssi2 npr1 were digested with EcoRV and resolved on an agarose gel. The size of each band in base pairs is listed at left.
(C) Comparison of PR1 expression in the leaves of 4-week-old soil-grown ssi2 plants that are homozygous for the wild-type FAD6 allele (ssi2), heterozygous for the fad6-1 mutant allele (ssi2 fad6/+), and homozygous for the fad6-1 mutant allele (ssi2 fad6). All RNAs were resolved on denaturing gels, transferred to Nytran Plus membranes, and probed for the indicated genes. Gel loading was monitored by photographing the ethidium bromide–stained gel (EtBr) before transferring the RNA to a Nytran Plus membrane.
(D) Morphological phenotype of 4-week-old soil-grown ssi2 plants that are homozygous for the wild-type FAD6 allele (ssi2), heterozygous for the fad6-1 mutant allele (ssi2 fad6/+), and homozygous for the fad6-1 mutant allele (ssi2 fad6). All plants were photographed from the same distance.
(E) Light microscopy of trypan blue–stained leaves of 4-week-old soil-grown ssi2 plants that are homozygous for the wild-type FAD6 allele (ssi2), heterozygous for the fad6-1 mutant allele (ssi2 fad6/+), and homozygous for the fad6-1 mutant allele (ssi2 fad6). All photographs were taken at the same magnification.