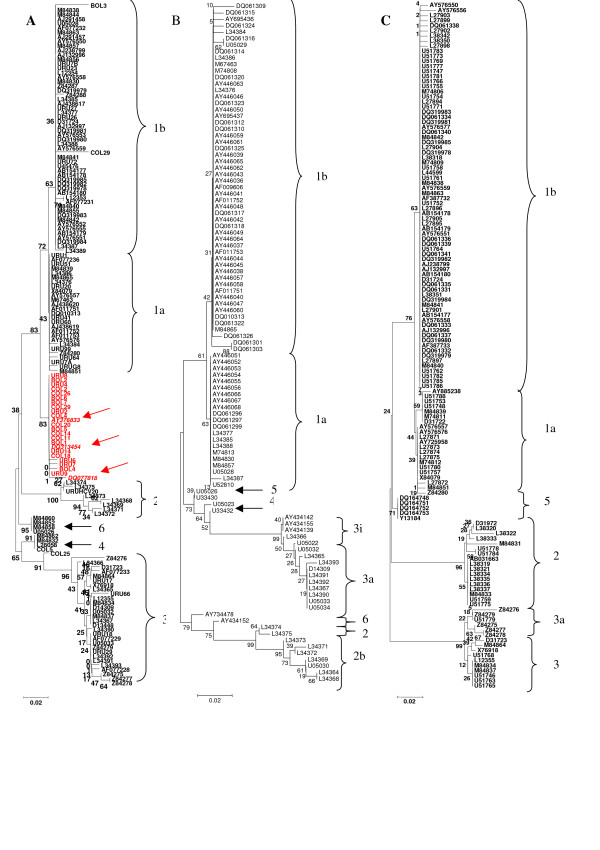
Figure 1.
Phylogenetic analysis of 5'NCR sequences of HCV strains. Strains in the trees are shown by their accession numbers for strains previously described and their genotypes are indicated at the right side of the figure. Bolivian, Colombian and Uruguayan strains are shown by name. Number at the branches show bootstrap values obtained after 1000 replications of bootstrap sampling. Bar at the bottom of the trees denotes distance. In (A) the phylogenetic tree for HCV strains isolated in South America is shown. Strains assigned to a newly genetic lineage in HCV type 1 cluster are shown in red. Argentinean strains [EMBL:DQ077818] (Schijman et al., unpublished data), [EMBL:DQ313454] and [EMBL:AY376833] (Gismondi et al. [8, 9] previously reported as a new genetic lineage inside type 1 strains are shown in italics and an arrows denote its position in the figure. Phylogeny for HCV strains isolated in North America and Europe are shown in (B), (C), respectively.