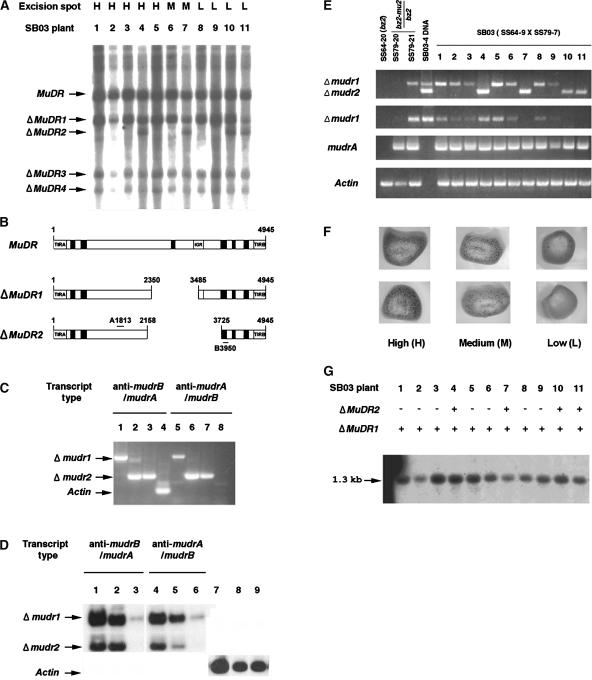
Figure 4.
Molecular and Structural Analyses of Two Germinally Deleted MuDR Elements and Their Transcripts.
(A) Detection of heritable deleted MuDR derivatives in sibling plants grown from SB03. Genomic DNA was prepared from these plants, digested with SstI, and blotted and probed with the A and B probe described in Figure 1B. Four deleted elements (ΔMuDR1 to ΔMuDR4) were found and are indicated with arrows. H, high; M, medium; L, low.
(B) Schemes of the intact MuDR and two ΔMuDR elements. Primers 3 and 12 shown in Figure 1A were used to amplify portions of ΔMuDR1 and ΔMuDR2. The amplified fragments were sequenced to confirm the structures.
(C) Directional RT-PCR cloning of antisense mudrB/mudrA (anti-mudrB/mudrA) and antisense mudrA/mudrB (anti-mudrA/mudrB) from total RNA of plants SB03-1 (lanes 1 and 5), SB03-4 (lanes 2 and 6), and SB03-11 (lanes 3 and 7). Single-stranded cDNA was obtained using either primer 12 (lanes 1, 2, and 3) or primer 3 (lanes 5, 6, and 7). After removal of template RNA and inactivation of the reverse transcriptase, cDNA was amplified using primer 3 or primer 12, as appropriate. Control amplification of sense actin transcripts (lane 4), but not antisense transcripts (lane 8), confirmed that the RT-PCR was directional in this experiment. Data show RT-PCR products fractionated on a 1% agarose gel and stained with ethidium bromide.
(D) Amplification of the Δmudr transcripts from several RNA sources. Total RNA was extracted from plant SB03-4 and fractionated into poly(A+) and poly(A−) RNA as described in Methods. Directional RT-PCR was performed using total RNA (lanes 1, 4, and 7), poly(A−) RNA (lanes 2, 5, and 8), or poly(A+) RNA (lanes 3, 6, and 9) as described in (C). The amplified RT-PCR products were fractionated on an agarose gel and detected using the A and B DNA probes after blotting on a nylon membrane. Sense actin transcripts were amplified as a control.
(E) Transcripts of ΔMuDR1 and ΔMuDR2 and expression levels of sense mudrA transcripts. Transcripts were amplified from total RNA of SB03 plants, from two plants in the family of the male parent (SS79-20 and SS79-21), and from a plant in the family of the female parent (SS64-20). Primers 3 and 12 were used for the amplification of both Δmudr1 and Δmudr2 transcripts (top gel). RT-PCR with primers 4 and 12 produces only Δmudr1 transcripts (second gel). Sense mudrA transcripts were amplified using primers 5 and 9 (third gel). Note that these primers can amplify two known mudrA transcripts, because intron 3 is spliced in only ∼80% of the transcripts (Hershberger et al., 1995); in this experiment, most transcripts appear to be spliced, because the smaller 490-bp product is visualized prominently by ethidium bromide staining. The Actin gel (bottom) indicates that RT-PCR amplification was performed as a control to determine if equal amounts of RNA were used in the experiment. Portions of the ΔMuDR1 and ΔMuDR2 elements were amplified by PCR from genomic DNA of plant SB03-4 to use for a size comparison with the RT-PCR products (top two gels only; lane 4). The PCR amplification was for 31 cycles.
(F) Representative kernels of the SB03 ear (SS64-3 × SS79-7) were classified as high, medium, or low spotted. Mu excision from a mutable bz2-mu2 allele restores a functional Bz2 gene, and this is a visual indicator of somatic Mutator activity, which varied from high (>150 spots on a 2 mm × 2 mm aleurone area of the kernels) to medium (10 to 150 spots) to low (<10 spots).
(G) Analysis of the methylation status at HinfI sites within the MURA binding sites in the TIRs of Mu1 and Mu2 elements. Genomic DNA extracted from SB03 individuals was digested with HinfI, and the resulting DNA gel blot was probed with a Mu1/Mu2-specific probe (Raizada and Walbot, 2000). In this family, there were no Mu2 elements; consequently, only the 1.3-kb fragment generated from Mu1 was detected.