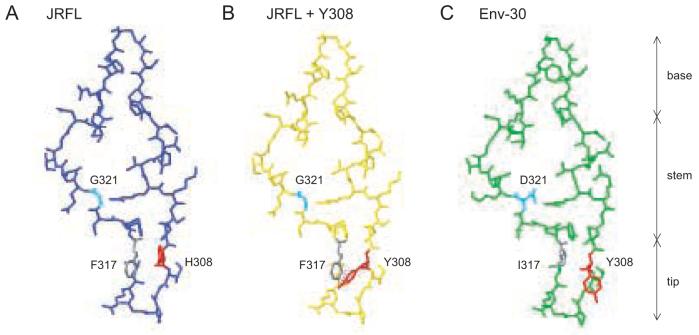
Figure 6. Structural modeling of Env V3 determinants contributing to broad coreceptor usage.
Swiss PDB viewer was used to model amino acid changes at positions 308, 317, and 321 on the V3 region of the JRFL gp120-CD4-X5 CD4i Ab crystal structure (2B4C) (Huang et al., 2005). JRFL V3 has a single amino acid change relative to the clade B consensus (N301Q). Red, position 308. Gray, position 317. Cyan, position 321. (A) V3 region of JRFL with H308, F317, and G321. (B) A Y308 change in JRFL results in steric clashing with F317 (dotted lines). (C) Changes Q301N, K305R, S306G, Y308H, F317I, G321D, E322K, I324V, Q328K, H330Y, and N332T were introduced in JRFL to produce a model of the V3 region of Env-30. Positions Y308, I317, and D321 are indicated.