Figure 1. Trans-splicing in S. cerevisiae can generate an RNA species comprising the 5’ end of U2 snRNA and the 3’ exon of the ACT1-CUP1 reporter.
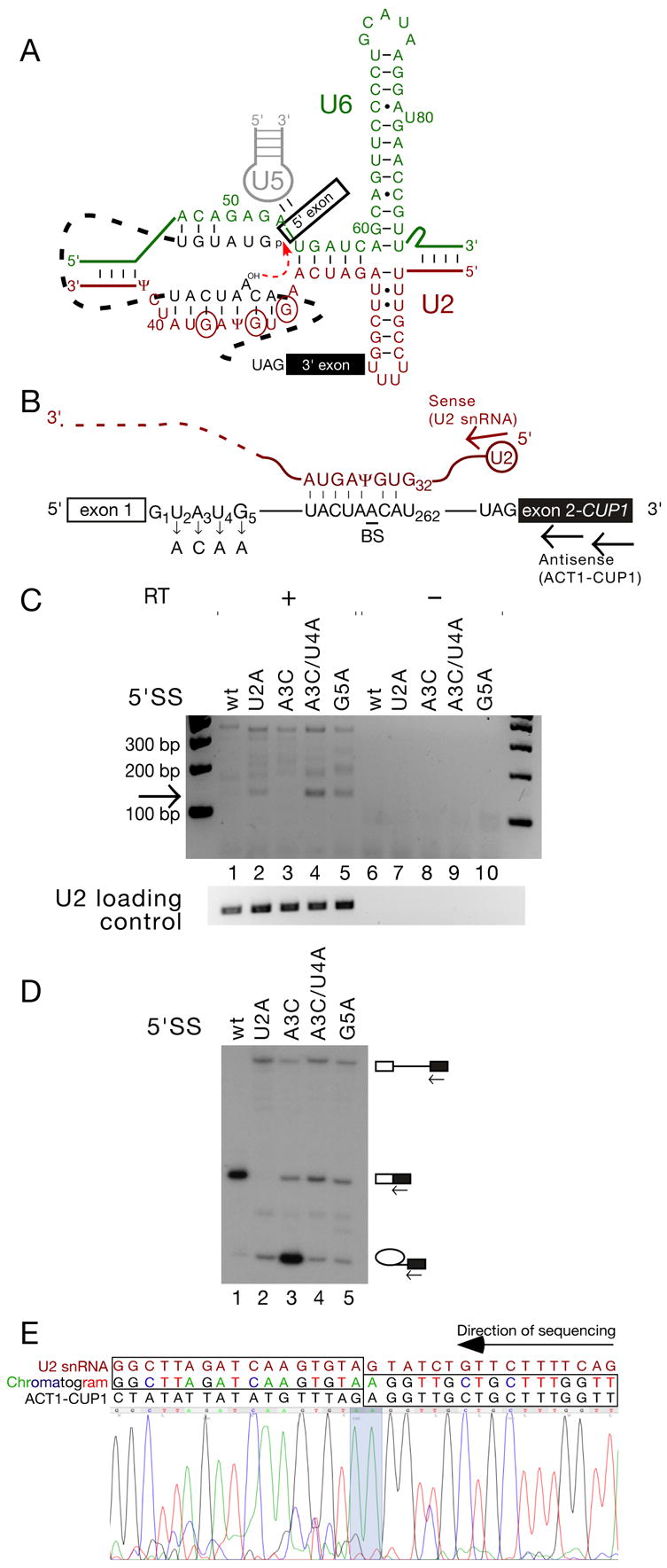
(A). Schematic of RNA-RNA interactions that contribute to the first step of splicing (modified from Konarska et al., 2006) with the initial G of each of the three 5’ splice site-like sequences in the BS-binding region of U2 snRNA indicated by circles.
(B). Schematic of ACT1-CUP1 reporter pre-mRNA and U2 snRNA, indicating the mutations used in panels C and D, and the location of the RT and PCR primers (arrows) used in panel C.
(C). RT-PCR using the primers indicated in Fig. 1B can amplify a product, of the size expected for a trans-splicing product generated using the BS-binding region of U2 as a 5’SS, from total RNA from S. cerevisiae Y04999 cells carrying reporters with 5’SS mutations as indicated.
(D). Primer extension analysis of RNA recovered from cells containing the ACT1-CUP1 reporters as indicated. Primer complimentary to the 3' exon was used to reveal levels of pre-mRNA, mRNA, and lariat intermediate. Strain Y04999 (Δdbr1) was used in order to accurately monitor the efficiency of the first step.
(E). The 130 bp RT-PCR product corresponds to trans-spliced U2-ACT1-CUP1. Reverse-complemented sequencing trace from the purified 130 bp product. U2 snRNA and ACT1-CUP1 sequence, as well as the chromatogram read, are indicated above the trace; concordance between chromatogram and gene sequence is indicated by boxes, and the splice junction is highlighted on the chromatogram trace.
