Abstract
GT-2 is a plant transcriptional activator that contains two separate, but similar, trihelix DNA-binding domains. GT-1 is similar to GT-2, although it contains only one of such domains. cDNAs that encode GT-2 were isolated from rice (OS-GT2) and Arabidopsis (AT-GT2). Evidence is presented for the existence of an Arabidopsis gene family that is structurally related to AT-GT2. Two members of this GT2-like family, AT-GTL1 and AT-GTL2, have been isolated and characterized. Their sequences suggest that they evolved by a recent gene duplication event. Both AT-GT2 and AT-GTL genes contain an intron in the amino-terminal trihelix motif, indicating that this DNA-binding domain resulted from exon shuffling. RNA gel blot analysis using AT-GTL1 as a probe revealed four transcripts in the aerial part of the plant. All mRNA levels were significantly higher in siliques, suggesting that this gene family may function in fruit and/or seed development. To date, DNA-binding proteins characterized by the trihelix motif have been described only in plants, and may therefore be involved in plant-specific processes. Our results show that in Arabidopsis thaliana, the trihelix motif is not restricted to the GT-1 and GT-2 DNA-binding proteins.
Keywords: duplication, evolution, trihelix motif
A vast number of genes encoding transcription factors have been identified in a wide variety of organisms and their characterization led to the recognition of common DNA-binding and/or protein interaction motifs on the basis of which a classification was made (1). Some classes of transcription factors have been identified in all kinds of higher organisms, whereas others appear to be restricted to a specific branch of the evolutionary tree. For example, members of the homeobox gene family are ubiquitously represented, whereas genes containing a homeobox fused to a leucine zipper motif appear specific for plants (HD-ZIP proteins; ref. 2). Another class of factors found uniquely in plant species contains the trihelix DNA-binding motif (3–5). Members of this family include GT-2 from rice and GT-1 from tobacco, orthologs of which were also characterized in Arabidopsis thaliana, where both are encoded by a single-copy gene (6, 7). GT-1 and GT-2 are nuclear factors that interact with light-responsive gene promoters and are homologous within their functionally defined DNA-binding domains. GT-2 differs structurally from GT-1 in that it has twin DNA-binding domains whereas GT-1 possesses only one trihelix DNA-binding motif (3–5, 8). Nuclear localization signals were identified for both GT-1 and GT-2, and the latter was shown to exhibit transcriptional activation activity in vivo (7, 9, 10). For GT-1, an oligomerization domain was identified suggesting a regulation of transcription in a hetero- or homodimeric form (7, 8).
Here we show that the Arabidopsis genome contains at least two genes that encode proteins homologous and structurally similar to AT-GT2, indicating that the trihelix DNA-binding motif may be more widespread among plant-regulatory proteins.
MATERIALS AND METHODS
Plant Material and Growth Conditions.
A. thaliana Heynh. ecotypes Columbia (Col-0) and Landsberg erecta were obtained from Lehle Seeds (Round Rock, TX). A set of 30 recombinant inbred lines generated from a cross between Landsberg erecta and Col-0 (11) was obtained from the Nottingham Arabidopsis Stock Centre.
Plants were grown on soil in a growth chamber at 22°C and 60% relative humidity, under white fluorescent light (75 μmol/m2·s) and long-day conditions (16 h light/8 h dark).
DNA Manipulations.
Standard methods were used for DNA manipulations, including the purification of plant, phage, and plasmid DNA, the preparation of Southern filters, the screening of λ phage libraries, and the enzymatic manipulation of cloned or genomic DNA (12). Filter hybridizations were conducted according to Church and Gilbert (13). Plant genomic DNA was prepared from 2-week-old light-grown Col-0 seedlings.
An Arabidopsis (Col-0) genomic library in λgem11 vector (a gift from Chris Somerville, Carnegie Institute of Washington, Stanford, CA) was screened with the EcoRI insert of a λgt11 cDNA clone (hereafter named R64). R64 was isolated in a screen for proteins binding to the promoter of the AT-ACS1 gene (14) and encodes a polypeptide with significant homology to the Arabidopsis GT-2 (J.S., unpublished data). Hybridizations were performed at 60°C, and washing was done at room temperature in a buffer of 2× standard saline citrate (SSC; 1× SSC = 0.15 M NaCl/0.015 M Na3-citrate, pH 7.0) and 0.1% SDS. Four clones hybridized to both HindIII/EcoRI fragments of the R64 cDNA (i.e., R16 and R31), encoding parts of the R64 protein that are homologous to the 5′ and 3′ GT-2 DNA-binding domains, respectively. These clones were further analyzed.
DNA Sequencing.
DNA from each of the four λ clones was digested with HindIII, BglII, BamHI, and EcoRI. Fragments were subcloned into pBS+ (Stratagene). For each digest, subclones hybridizing to the R64 cDNA were sequenced by using an automated sequencer (Applied Biosystems) and directed oligonucleotide primers. Gene structure was confirmed by PCR by using gene-specific primers on Col-0 genomic DNA. Data analysis was performed by using the Genetic Computer Group (Madison, WI) sequence analysis software (15). For subclones derived from three out of four originally hybridizing λ clones, homology to R64 was confirmed on the sequence level; the fourth clone, however, did not contain any significant homology. This clone (pJSRL6) was discarded as a false positive. The protein sequence alignment was displayed using seqvu 1.0 (The Garvan Institute of Medical Research, Sydney, Australia).
Gene Mapping.
AT-GT2 and AT-GTL2 were positioned on the Arabidopsis genetic map by restriction fragment length polymorphism (RFLP) segregation analysis by using recombinant inbred lines as described by Lister and Dean (11). Chromosome positions are shown relative to flanking markers.
RNA Isolation and Gel Blot Analysis.
Total RNA was extracted from young (unexpanded) and old (fully expanded) leaves, stems, flowers, siliques, and roots of 5-week-old Col-0 plants according to Logemann et al. (16). For RNA gel blot analysis, samples of 30 μg of total RNA were electrophoresed in a 1.5% agarose-formaldehyde gel and transferred to a nylon filter (Hybond-N; Amersham). To estimate whether equal amounts were loaded, the RNA was visualized in the gel by adding 1 μl of ethidium bromide (stock concentration of 0.5 mg/ml) to each sample. For detection of GTL transcripts, a 32P-labeled riboprobe was synthesized from SpeI/XbaI-linearized pJSR16 (containing the HindIII/EcoRI fragment at the 5′ end of the R64 cDNA and encoding the amino-terminal DNA-binding domain) by using T3 polymerase (Riboprobe Gemini II core system; Promega). Hybridization, washing, and autoradiography were done as described by Kurepa et al. (17). The sizes of the hybridizing transcripts were estimated relative to known RNA standards (GIBCO/BRL).
RESULTS
Isolation of Genes Homologous or Identical to Arabidopsis GT-2.
In a study aimed at identifying genes whose products bind to the promoter of the Arabidopsis AT-ACS1 gene, a cDNA was isolated encoding a protein with strong homology to the DNA-binding domain of both rice and Arabidopsis GT-2 (J.S., unpublished data). This cDNA (named R64) was used to screen a genomic library resulting in the isolation of three genomic clones (see Materials and Methods). Within its coding region one of these genes was 100% identical to the R64 cDNA and was named AT-GTL1 (acronym for A. thaliana GT2-like-1). A second gene, AT-GTL2 was identical to AT-GTL1, except for a truncation at the carboxyl-terminal part of the coding region at position 1777 (numbering from the start codon and including the intron sequence), resulting in a shorter predicted protein (474 amino acids instead of 594 for AT-GTL1). The third clone hybridized only weakly to the R64 cDNA and was, except for a single intron, identical to a cDNA encoding Arabidopsis GT-2 (6).
Analysis of the Predicted ORFs.
Fig. 1A shows the homology between the predicted AT-GTL1 protein and GT2 factors from rice (OS-GT2) and Arabidopsis (AT-GT2). All three proteins shared a high degree of similarity in three separate domains, previously identified as the amino-terminal DNA-binding motif, the central domain, and the carboxyl-terminal DNA-binding motif (6). In addition, various other regions with a significant similarity could be discerned. AT-GTL1 and OS-GT2 shared some identity in their glycine-rich amino-terminal part, whereas AT-GTL1 and AT-GT2 had a higher level of identity between the amino-terminal DNA-binding motif and the central domain. As a result, it was difficult to determine whether AT-GTL1 was more related to the Arabidopsis than to the rice GT-2. The overall percentage of similarity between AT-GTL1 and AT-GT2, respectively OS-GT2, was almost the same (i.e., 37% for AT-GT2 and 36% for OS-GT2). Based on the similarity within the three highly conserved domains, a simple and straightforward relation was also absent (Fig. 1B). When considering the amino-terminal and central domains, the AT-GTL1 protein was more related to rice than to Arabidopsis GT-2. In contrast, in its carboxyl-terminal domain AT-GTL1 shared the same overall similarity to both GT-2 proteins. The predicted secondary structure of both DNA-binding motifs of AT-GTL1 corresponds with the trihelix structure that was found in both rice and Arabidopsis GT-2 DNA-binding domains (data not shown).
Figure 1.

Similarity between proteins with twin trihelix DNA-binding motifs. (A) Amino acid sequence comparison between the predicted GTL1 protein (AT-GTL1) and GT-2 from Arabidopsis (AT-GT2) and rice (OS-GT2). The analysis was performed by using the pileup program of the General Computer Group package, and sequence identity was displayed using seqvu 1.0. Gray boxed sections represent the amino-terminal and carboxyl-terminal DNA-binding motifs whereas the solid boxed amino acids belong to the conserved central domain. (B) Schematic representation of the sequence comparison depicted in A. Gray boxes represent the amino-terminal (NT) and carboxyl-terminal (CT) DNA-binding motifs. The conserved central domain (CD) is indicated in black. Length of proteins is displayed on the right-hand side. The scheme of the AT-GTL1 protein is presented twice to enable a clear display of the amino acid identity levels.
As in AT-GT2 and OS-GT2, the AT-GTL1 protein contained glutamine and proline-rich regions in between the two DNA-binding motifs, as well as a stretch of acidic amino acids in the carboxyl-terminal part (Fig. 1A). In addition, both trihelix motifs also contained bipartite nuclear localization sequences, identical to those found in rice and Arabidopsis GT-2 (9, 18).
Gene Organization.
The AT-GT2, AT-GTL1, and AT-GTL2 genes all contained a single intron in the amino-terminal DNA-binding domain with intron/exon junctions in the same position relative to the conserved residues of this motif (Fig. 2). The intron in AT-GT1 was located between amino acids 96 and 97, whereas in AT-GT2 it was positioned between residues 75 and 76. The R64 cDNA was, except for the intron, identical to AT-GTL1 and was renamed AT-GTL1 cDNA. Comparison of the intron, promoter, and 3′ noncoding sequences of AT-GT2 and AT-GTL1 did not reveal any significant similarities (data not shown). The AT-GTL2 gene was truncated relative to AT-GTL1, and this truncation was located in the carboxyl-terminal DNA-binding domain in a position similar to the intron in the amino-terminal domain (Fig. 2).
Figure 2.
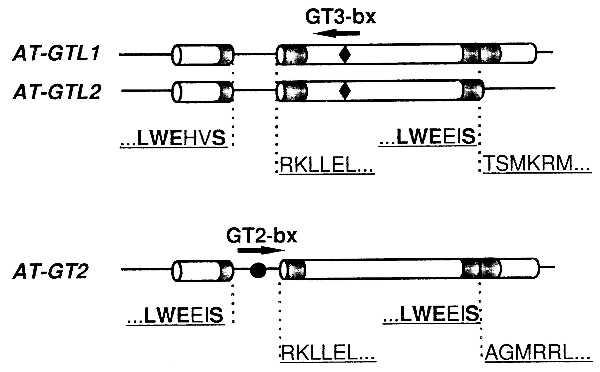
Intron/exon structure of Arabidopsis GTL and GT-2 genes relative to amino acid sequences of the trihelix motifs (indicated in gray). Amino acids conserved among all trihelix motifs are highlighted in bold. AT-GT2 contains a GT2-binding site (GT2 bx, indicated by •) in its intron. AT-GTL1 and AT-GTL2 have an inverted GT3 target site (GT3 bx, indicated by ⧫) in their second exon.
All three genes were analyzed for GT box motifs (3). The AT-GT2 gene contained a GT2-binding motif in the intron, whereas AT-GTL1 and AT-GTL2 had an inverted GT3-box in the second exon (Fig. 2).
Fig. 3A shows the result of a high-stringency genomic DNA gel blot experiment in which BamHI-digested DNA was probed with the HindIII/EcoRI insert of pJSR31 (an AT-GTL1 cDNA subclone containing the carboxyl-terminal trihelix motif, not digested by BamHI; Fig. 3B). A pattern of at least three bands was observed indicating the presence of at least one more sequence in the Arabidopsis genome highly similar to AT-GTL1 and AT-GTL2 (Fig. 3A). The same blot was probed with pJS75RG2, a BglII subclone (1723 bp insert) that contained part of the AT-GT2 gene including the carboxyl-terminal trihelix motif, and with pJSRL6, a clone that contained a single-copy sequence defining an RFLP marker located in the middle of chromosome 1 (see below). Both the inserts of pJS75RG2 and pJSRL6 had no internal BamHI site and hybridized to single BamHI bands in the genomic DNA gel blot.
Figure 3.
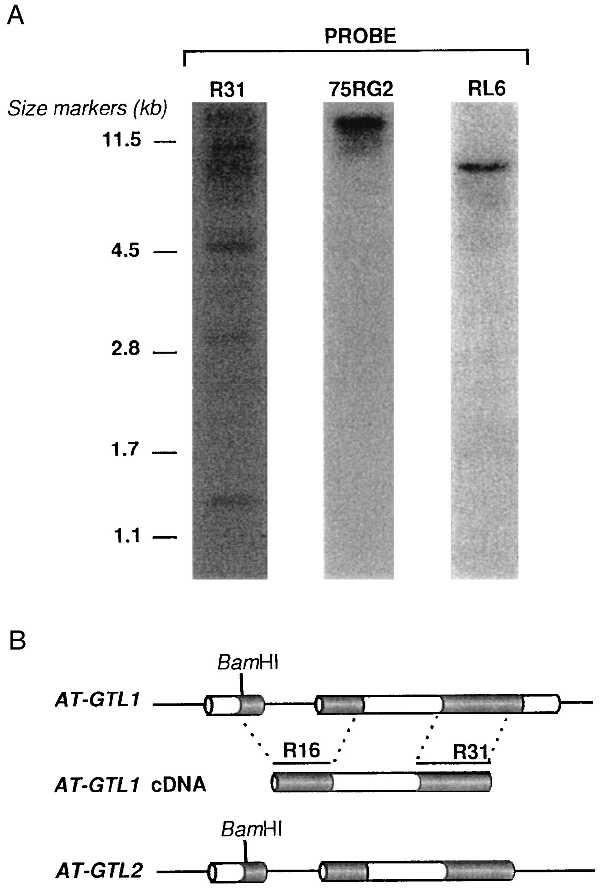
GTL genes in the Arabidopsis genome. (A) DNA gel blot with BamHI-digested Arabidopsis genomic DNA after hybridization with the insert of pJSR31 that encodes the carboxyl-terminal trihelix motif of AT-GTL1 (R31), after hybridization with pJS75RG2 that encodes a part of AT-GT2 (75RG2), and after hybridization with pJSRL6 (unrelated to either one) (RL6). Size markers identify some of the bands of PstI-digested λ DNA. (B) Schematic representation of the AT-GTL1 and AT-GTL2 genes and the AT-GTL1 cDNA. Shown are the exons (boxes), introns and untranslated sequences (lines), and the single BamHI site in the first exon. R16 and R31 are HindIII/EcoRI fragments of the AT-GTL1 cDNA that contain the amino-terminal or carboxyl-terminal DNA-binding motif, respectively. The R31 fragment was used in the genomic DNA gel blot analysis (Fig. 3A). A subclone containing R16 was used in RNA gel blot analysis (see Fig. 5A).
Chromosomal Location of the AT-GT2 and AT-GTL2 Genes.
To determine map positions, subclones of each gene were used to detect RFLPs in DNA prepared from the Col-0 and Ler ecotypes. For AT-GT2, an RFLP was detected between Ler and Col-0, by hybridizing pJS75RG2 (containing the central BglII fragment of the gene) to BclI-digested DNA. For AT-GTL2 an RFLP was found by hybridizing pJS69RG7 (containing a BglII-BamHI fragment covering the partial carboxyl-terminal DNA-binding domain and the 3′ noncoding region) to HincII-digested DNA. The AT-GT2 and AT-GTL2 genes were both located on chromosome 1 (Fig. 4). AT-GT2 maps 1 cm from the hypocotyl mutant botero1 (H. Höfte, personal communication). The false-positive pJSRL6 (see Materials and Methods) was also mapped by using a HincII polymorphism and is located 2 cM from the m213 marker on chromosome 1 (19).
Figure 4.
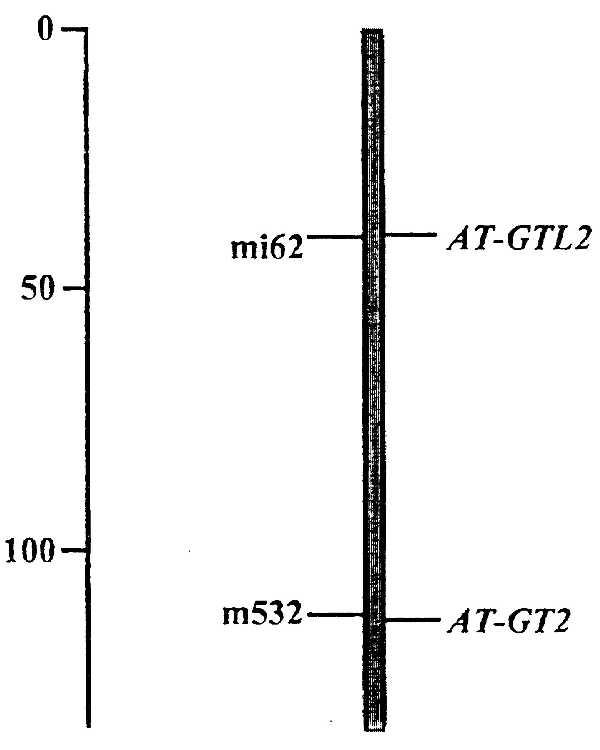
Chromosomal location of AT-GT2 and AT-GTL2. Both genes map to chromosome 1. Positions are shown relative to flanking RFLP markers: mi62 (20) for AT-GTL2 (distance 0.5 cM), and m532 (19) for AT-GT2 (distance 0.2 cM). Numbers on the left-hand side indicate distances in cM.
Expression Pattern of GTL Genes.
To detect transcripts of the AT-GTL1 and AT-GTL2 genes, RNA gel blot analysis was performed by using pJSR16 (an AT-GTL1 cDNA subclone encoding the amino-terminal trihelix motif; see Fig. 3B). Probes containing the less conserved region flanked by the DNA-binding domains could not be used for this purpose because they hybridized to a large number of bands in genomic DNA gel blot experiments (data not shown). In leaves, stems, flowers, and siliques, a set of four transcripts was detected (2.9, 2.0, 1.8, and 1.4 kb), all of which were induced in fruits (Fig. 5A). Previously, a 2.1-kb mRNA was detected by using the Arabidopsis GT-2 cDNA as a probe (6). Low transcript levels were detected in young and old leaves, stems, and flowers, whereas no signal was observed in roots.
Figure 5.
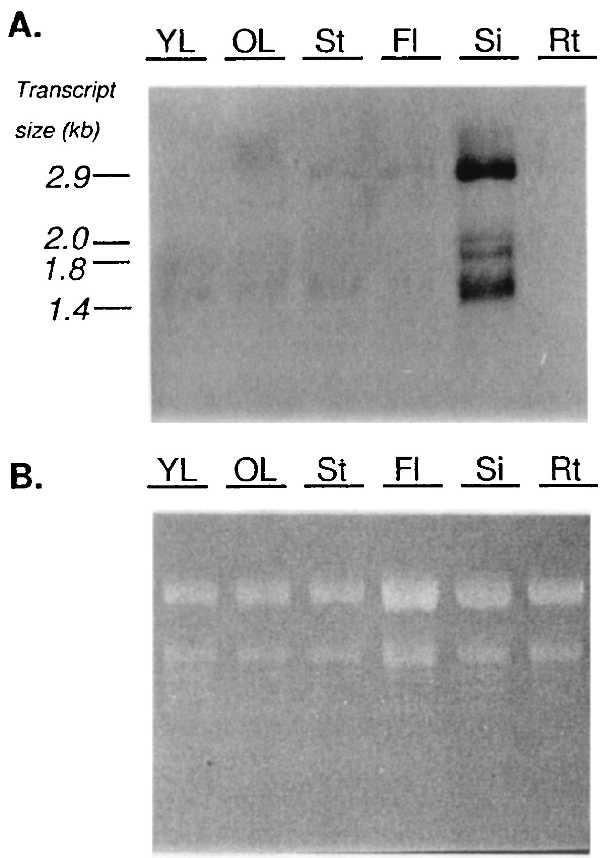
RNA gel blot analysis of the expression of GTL genes in Arabidopsis organs. Total RNA was isolated from young (YL) and old (OL) leaves, stems (St), flowers (Fl), siliques (Si), and roots (Rt) of 5-week-old Col-0 plants. The RNA gel blot was probed with 32P-labeled antisense R16 (see Fig. 3B). The autoradiogram (5 days of exposure) is shown in A and the ethidium bromide-stained gel is shown in B as a control for equal loading. Length of transcripts (in kb) is given on the left.
DISCUSSION
The DNA-binding proteins GT-1 and GT-2 define a class of factors so far uniquely found in plant species (10). This indicates that the trihelix DNA-binding motif emerged after the divergence of plants and animals, and suggests that this kind of proteins regulate plant-specific processes. Both GT-1 and GT-2 are expected to be involved in the transcriptional control of light-regulated genes (3–5). Here we show that in Arabidopsis thaliana the structure of twin trihelix DNA-binding domains is not restricted to AT-GT2. Although Arabidopsis has one of the smallest genomes among the angiosperms (21, 22), it is estimated that >15% of the Arabidopsis genes may be encoded by multiple loci (23). The AT-GTL1 and AT-GTL2 genes are almost identical and probably result from a recent duplication event. High-stringency DNA gel blot experiments have also shown that at least one more highly similar gene may exist in the Arabidopsis genome. In addition, RNA gel blot analysis using part of the GTL1 cDNA as a probe revealed four transcripts that are all induced in siliques (Fig. 5). Although these transcripts may represent different AT-GTL genes, we cannot exclude the possibility that they originated from alternative transcription initiation sites or from different transcript processing. Cloning of the remaining AT-GTL sequences and analysis with gene-specific probes will be necessary to answer this question.
On low-stringency genomic DNA gel blots it was not possible to detect the AT-GT2 gene by using probes for the AT-GTL1-coding region and vice versa (data not shown). Nevertheless, upon screening of a genomic library, AT-GT2 was isolated by using parts of the AT-GTL1 cDNA (encoding the DNA-binding domains) as a probe. It remains possible that similar genes exist that have diverged from both AT-GT2 and AT-GTL genes, and that have not been detected under our screening conditions.
The exon/intron structure of AT-GT2 and AT-GTL1 is identical. The amino-terminal trihelix is interrupted in the second helix, suggesting that this DNA-binding domain is the result of exon shuffling (Fig. 2). In contrast, the carboxyl-terminal domain is continuous in AT-GT2 and AT-GTL1 whereas it is truncated in AT-GTL2 in a position identical to the intron insertion site in the amino-terminal domain. Sequencing of AT-GTL2 farther downstream (0.5 kb) did not reveal a third exon that continues the trihelix motif. In addition, PCR on the AT-GTL2 genomic λ clone, using oligonucleotide primers covering a putative intron, did not amplify any fragment (data not shown). It remains possible that a large intron difficult to overspan by a classical PCR divides the carboxyl-terminal trihelix motif of AT-GTL2.
The structure of AT-GTL2 suggests that twin trihelix factors originated from the second exon that was duplicated twice, thus generating the second helix of both DNA-binding domains (Fig. 6). Subsequently, the ancestral gene could have evolved into an AT-GT2-like structure by loss of the intron in the carboxyl-terminal trihelix, and by divergence of the 5′ and 3′ sequences. This hypothesis, however, is contradicted by the sequence identity of AT-GTL1 and AT-GTL2. If AT-GTL2 reflects an ancestral gene structure, then AT-GTL1 should have diverged from AT-GTL2. Alternatively, it remains possible that AT-GTL1 and AT-GTL2 have coevolved as part of a gene cluster where sequence identity was maintained by crossover fixation (24). However, as the map position of AT-GTL1 remains to be determined, we do not know whether AT-GTL1 is located in the vicinity of AT-GTL2 in the Arabidopsis genome. Previous studies have shown that the majority of duplicated copies of a given sequence are not tightly linked with one another and are even often located on different chromosomes (23).
Figure 6.
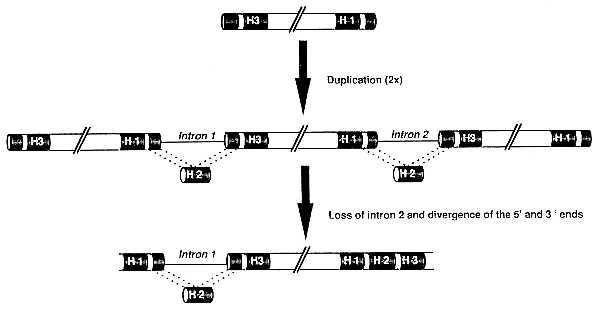
Model for the evolution of twin trihelix DNA-binding proteins. H1, H2, and H3 represent the helices of the trihelix DNA-binding motif (indicated in black). An ancestral exon containing the third (H3) and first (H1) helix is duplicated twice, resulting (after intron splicing) in the formation of the second helix (H2) and DNA-binding activity of two trihelix motifs. After loss of the second intron and divergence of 5′ and 3′ sequences, a gene structurally similar to AT-GT2 and AT-GTL1 emerges.
No significant homologies were found between the noncoding regions of AT-GT2 and AT-GTL1. The AT-GT2 gene contains a GT2-binding site in its intron suggesting that AT-GT2 autoregulates its expression. Recently, evidence was found for autoregulation of transcription factors in lower as well as higher organisms (25–28). This may be a mechanism by which a cell can rapidly respond to the need for enhanced target gene expression in response to an external stimulus. Alternatively, an almost immediate response may be controlled by primary stimulus—response factors that are posttranslationally regulated. Evidence for both transcriptional and posttranslational control emerge for instance in the case of response to the plant hormone abscisic acid (29). AT-GTL1 and AT-GTL2 have an inverted GT3 box in their second exon. However, its location and orientation suggest that this sequence probably has no functional role in the expression of either one of these genes. Although characterized by DNA-binding domains highly similar to AT-GT2, it is not known whether AT-GTL1 and AT-GTL2 have the same binding site specificity, nor whether they regulate the expression of light-regulated genes, as is the case for GT-2.
Several classes of DNA-binding proteins have been identified in Arabidopsis, and mutations in some of these factors have been linked to phenotypical alterations (30–35). Although expected to function in the light-regulated control of gene expression, no phenotypes have been associated with mutations in genes containing the trihelix DNA-binding domain. One reason could be that these genes may be regulating processes unidentified or uncharacterized to date. Alternatively, the close resemblance between AT-GT2, AT-GTL1, and AT-GTL2 may reflect functional redundancy and, thus, would mask loss of function mutations. We have constructed transgenic Arabidopsis lines underexpressing the AT-GTL1-coding sequence (J.S., unpublished data). This type of dominant mutations should allow us to determine whether trihelix factors play an essential role in plant growth and development.
Acknowledgments
We thank the Nottingham Arabidopsis stock center for the recombinant inbred lines, Claire Lister for assistance in mapping the genes, Chris Somerville for providing the Columbia genomic library, Luc Van Wiemeersch for help in sequence analysis, and Martine De Cock for help finalizing the manuscript. This work was supported by the Vlaams Actieprogramma Biotechnologie (ETC 002) and the Fund for Scientific Research (Flanders) (Krediet aan Navorsers 1993–1996). D.V.D.S. is a Research Associate of the Fund for Scientific Research (Flanders).
ABBREVIATION
- RFLP
restriction fragment length polymorphism
Footnotes
References
- 1.Pabo C O, Sauer R T. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 2.Ruberti I, Sessa G, Lucchetti S, Morelli G. EMBO J. 1991;10:1787–1791. doi: 10.1002/j.1460-2075.1991.tb07703.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dehesh K, Hung H, Tepperman J M, Quail P H. EMBO J. 1992;11:4131–4144. doi: 10.1002/j.1460-2075.1992.tb05506.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilmartin P M, Memelink J, Hiratsuka K, Kay S A, Chua N-H. Plant Cell. 1992;4:839–849. doi: 10.1105/tpc.4.7.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perisic O, Lam E. Plant Cell. 1992;4:831–838. doi: 10.1105/tpc.4.7.831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn R M, Caspar T, Dehesh K, Quail P H. Plant Mol Biol. 1993;23:337–348. doi: 10.1007/BF00029009. [DOI] [PubMed] [Google Scholar]
- 7.Hiratsuka K, Wu X, Fukuzawa H, Chua N-H. Plant Cell. 1994;6:1805–1813. doi: 10.1105/tpc.6.12.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lam E. Mol Cell Biol. 1995;15:1014–1020. doi: 10.1128/mcb.15.2.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dehesh K, Smith L G, Tepperman J M, Quail P H. Plant J. 1995;8:25–36. doi: 10.1046/j.1365-313x.1995.08010025.x. [DOI] [PubMed] [Google Scholar]
- 10.Ni M, Dehesh K, Tepperman J M, Quail P H. Plant Cell. 1996;8:1041–1059. doi: 10.1105/tpc.8.6.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lister C, Dean C. Plant J. 1993;4:745–750. [Google Scholar]
- 12.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology 1987–1988. New York: Greene Publishing Associates & Wiley-Interscience; 1987. [Google Scholar]
- 13.Church G M, Gilbert W. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Der Straeten D, Rodrigues-Pousada R A, Villarroel R, Hanley S, Goodman H M, Van Montagu M. Proc Natl Acad Sci USA. 1992;89:9969–9973. doi: 10.1073/pnas.89.20.9969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Logemann J, Schell J, Willmitzer L. Anal Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 17.Kurepa J, Bueno P, Kampfenkel K, Van Montagu M, Van den Bulcke M, Inzé D. Plant Physiol Biochem. 1997;35:467–474. [Google Scholar]
- 18.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 19.Chang C, Bowman J L, DeJohn A W, Lander E S, Meyerowitz E M. Proc Natl Acad Sci USA. 1988;85:6856–6860. doi: 10.1073/pnas.85.18.6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y-G, Mitsukawa N, Lister C, Dean C, Whittier R F. Plant J. 1996;10:733–736. doi: 10.1046/j.1365-313x.1996.10040733.x. [DOI] [PubMed] [Google Scholar]
- 21.Leutwiler L S, Hough-Evans B R, Meyerowitz E M. Mol Gen Genet. 1984;194:15–23. [Google Scholar]
- 22.Arumuganathan K, Earle E D. Plant Mol Biol Rep. 1991;9:208–218. [Google Scholar]
- 23.McGrath J M, Jancso M M, Pichersky E. Theor Appl Genet. 1993;86:880–888. doi: 10.1007/BF00212616. [DOI] [PubMed] [Google Scholar]
- 24.Smith G P. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- 25.Liptay S, Schmid R M, Nabel E G, Nabel G J. Mol Cell Biol. 1994;14:7695–7703. doi: 10.1128/mcb.14.12.7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delahodde A, Delaveau T, Jacq C. Mol Cell Biol. 1995;15:4043–4051. doi: 10.1128/mcb.15.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou P, Thiele D J. Genes Dev. 1993;7:1824–1835. doi: 10.1101/gad.7.9.1824. [DOI] [PubMed] [Google Scholar]
- 28.Sakamoto A, Omirulleh S, Nakayama T, Iwabuchi M. Plant Cell Physiol. 1996;37:557–562. doi: 10.1093/oxfordjournals.pcp.a028980. [DOI] [PubMed] [Google Scholar]
- 29.Shinozaki K, Yamaguchi-Shinozaki K. Plant Physiol. 1997;115:327–334. doi: 10.1104/pp.115.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yanofsky M F, Ma H, Bowman J L, Drews G N, Feldmann K A, Meyerowitz E M. Nature (London) 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- 31.Oppenheimer D G, Herman PL, Sivakumaran S, Esch J, Marks M D. Cell. 1991;67:483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 32.Deng X-W, Matsui M, Wei N, Wagner D, Chu A M, Feldmann K A, Quail P H. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 33.Schena M, Lloyd A M, Davis R W. Genes Dev. 1993;7:367–379. doi: 10.1101/gad.7.3.367. [DOI] [PubMed] [Google Scholar]
- 34.Jofuku K D, den Boer B G W, Van Montagu M, Okamuro J K. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weigel D. Plant Cell. 1995;7:388–389. doi: 10.1105/tpc.7.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]


