Abstract
Transformation of chicken fibroblasts in vitro by Rous Sarcoma Virus represents a model of cancer in which a single oncogene, viral src, uniformly and rapidly transforms primary cells in culture. We experimentally surveyed the transcriptional program affected by Rous Sarcoma Virus (RSV) in primary culture of chicken embryo fibroblasts. As a control, we used cells infected with non-transforming RSV mutant td106, in which the src gene was deleted. Using Affymetrix GeneChipR Chicken Genome Arrays, we report 811 genes that were modulated more than 2.5 fold in the virus transformed cells. Among these, 409 genes were induced and 402 genes were repressed by viral src. From the repertoire of modulated genes, we selected 20 genes that were robustly changed. We then validated and quantified the transcriptional changes of most of the 20 selected genes by real-time PCR. The set of strongly induced genes contains vasoactive intestinal polypeptide, MAP kinase phosphatase 2 and follistatin, among others. The set of strongly repressed genes contains TGF beta 3, TGF beta-induced gene, and deiodinase. The function of several robustly modulated genes sheds new light on the molecular mechanism of oncogenic transformation.
Keywords: avian sarcoma, Schmidt-Ruppin strain of RSV, src oncogene, protein-tyrosine kinase, vasoactive intestinal polypeptide, MAP kinase phosphatase 2, follistatin
Introduction
Transformation of primary chicken fibroblasts (CEFs) in vitro by Rous sarcoma virus (RSV) continues to represent an important experimental model of cancer (Martin, 2004). The transforming gene of RSV, the viral src oncogene (v-src), is an activated and over-expressed protein-tyrosine kinase responsible for a number of molecular events that govern rapid and dramatic phenotypic changes observed in transformed host cells (Jove and Hanafusa, 1987; Blume-Jensen and Hunter, 2001; Darnell, 2002; Frame, 2002). Transformation-specific changes in cell morphology, proliferation and anchorage independence of growth are accompanied by changes in gene expression. In the past, a number of laboratories reported identification of individual genes that were either induced or repressed by the v-src oncogene at the level of transcription (Howard et al., 1978; Hendricks and Weintraub, 1981; Fagan et al., 1981; Sugano et al., 1987; Bedard et al., 1987; Jahner and Hunter, 1991; Dehbi and Bedard, 1992; Herault et al., 1992; Frankfort and Gelman, 1995). With the recent completion of the chicken genome sequence (Wallis et al., 2004; Hillier et al., 2004) and the subsequent availability of Affymetrix GeneChipR Chicken Genome Arrays that represent the full complement of the genome, we experimentally surveyed the transcriptional program affected by RSV transformation in the syngeneic system of Schmidt-Ruppin strain of RSV (RSV-SR) and primary culture of CEFs. As a control, we used CEFs infected with non-transforming RSV (td106) derived from the Schmidt-Ruppin strain of RSV in which the src gene was deleted (Wang et al., 1984). Thus, the isogenic pair of viruses differed only in the src gene, but not in the replicative genes, allowing one to score for transcripts relevant for src transformation. The rationale for the global analysis of transcriptional changes caused by v-src was to provide better understanding of the mechanism of transformation, taking advantage of a cancer model in which a single oncogene uniformly and rapidly transforms primary cells in culture (Brugge and Erikson, 1977).
Using GeneChipR Chicken Genome Arrays that cover 32,773 transcripts corresponding to 28,418 predicted genes from the Ensembl annotation of the complete genome, we report 409 genes that are induced and 402 genes that are repressed at the level of transcription by v-src. From the repertoire of modulated genes, we selected 20 genes (10 induced and 10 repressed) that were robustly changed by v-src in the screen of Affymetrix DNA arrays. We then validated and quantified the transcriptional changes of most of the 20 selected genes by real-time PCR using array-independent sets of primer oligos and new preparations of RNA. The function of several robustly induced and repressed genes shed new light on the molecular mechanism of oncogenic transformation.
Results and Discussion
Identification of v-src-modulated genes
Analysis of GeneChipR Chicken Genome Arrays probed with RSV-SR wild type and RSV-SR-td106 mutant derived probes revealed that circa 10% of transcripts were changed. From 28,418 chicken genes investigated in the screen, 17,868 were detected at least once in RSV-SR or RSV-SR-td106 mutant screens. Among the detected genes, 1602 were induced and 1386 were repressed in v-src transformed cells. These changes were considered significant as they had a p-value of 0.05 or less. When a filter of two moderately stringent criteria was applied to the data, namely that the changes had to be more than 2.5 fold (linear, in either direction) and the p-value had to be less than 0.05, the number of modulated transcripts decreased by more than 3 fold to 811. Among the selected transcripts, 409 were induced and 402 were repressed by v-src (see Figure A representing a heat map of hierarchical clustering and Table A listing all 811 modulated sequences, in Supplementary Material). This set of data contained several genes that had been previously documented as v-src-modulated genes: urokinase-type plasminogen activator, gene for glucose transporter protein, and nephroblastoma overexpressed gene (Leslie et al., 1990; White et al., 1991; Scholz et al., 1996).
Validating selected transcripts by RT-PCR
We chose 20 robustly modulated genes from the “stringent set” of 811 modulated transcripts for further analysis. Among these 20 genes, 10 were induced and 10 repressed (Table 1). We isolated new RNAs from independent RSV-SR transformed and RSV-SR-td106 infected CEFs and generated the array-independent set of primer oligos to validate and quantify the changes by another technique, real-time PCR (RT-PCR). Using a housekeeping transcript, GAPDH, as a control for quantification, and documenting the quality of each of the amplified products by melting curve analysis, we confirmed most (17 out of 20) of the transcriptional changes identified in the array screen. The changes scored by the GeneChipR Chicken Genome Array screen were in most cases larger than the changes measured by RT-PCR, even when the same RNA was used for the assays. This is often the case because of the intrinsic difficulty in setting the right background for the array analysis, especially with low signals from un-induced transcripts (personal communication from T. Hunter).
TABLE 1.
TRANSCRIPTS MODULATED IN RSV TRANSFORMED CHICKEN CELLS
| RSV (V-SRC) INDUCED GENES$ | |||||
| GENE SYMBOL | ACCESSION# | MW | GENE NAME | ARRAY(n=3) | RT-PCR(n=2) |
| PLAU | NM_205443 | 49 | plasminogen activator, urokinase | 196 ± 69 | 42 ± 11 |
| VIP | NM_205366 | 22 | vasoactive intestinal peptide | 124 ± 60 | 54 ± 0.5 |
| LOC423497 | XM_421400 | 353 | *AHNAK 1 & 2 related protein | 26 ± 5 | 8 ± 5 |
| LOC421581 | XM_419619 | 20 | *EMT organic cation transporter 3 | 18 ± 15 | N.C. |
| LOC423769 | XM_421641 | 65 | *arachidonate 5-lipoxygenase | 13 ± 6 | 7 ± 3.5 |
| LOC421692 | XM_419729 | 86 | *cAMP-specific phosphodiesterase 7B | 10 ± 5 | 10 ± 1.5 |
| FST | NM_205200 | 38 | follistatin | 9 ± 5 | 4 ± 0.5 |
| LOC423890 | XM_421754 | 75 | *dual specificity phosphatase 5 | 7 ± 4 | 7 ± 1.5 |
| NES | NM_205033 | 202 | Nestin | 7 ± 1 | 1 ± 0.2 |
| MKP-2 | NM_204838 | 41 | MAP kinase phosphatase 2 | 6 ± 2 | 26 ± 15 |
| RSV (V-SRC) REPRESSED GENES | |||||
| NPC2 | NM_001004393 | 21 | stathmin-like 3 | 45 ± 52 | 2 ± 0.2 |
| NOV | NM_205268 | 38 | nephroblastoma-overexpressed gene | 42 ± 39 | 18 ± 15 |
| TGFBI | NM_205036 | 74 | TGF beta-induced protein, 68 kDa | 28 ± 31 | 7 ± 4 |
| LOC418180 | XM_416411 | 23 | *RAB, estrogen-regulated growth inhibitor | 18 ± 8 | 2 ± 0.3 |
| LOC428711 | XM_426269 | 61 | four and a half LIM domains 1 protein | 15 ± 1 | 3 ± 0.5 |
| DIO2 | NM_204114 | 31 | deiodinase | 15 ± 6 | 5 ± 1 |
| C1QTNF3 | XP_425002 | 34 | C1q and TNF-related protein 3 | 13 ± 6 | 3 ± 0.5 |
| CPEB1 | XM_413713 | 80 | cytoplasmic polyadenylation element BP1 | 8 ± 3 | 0.3 ± 0.1 |
| TFPI | XM_421849 | 35 | tissue factor pathway inhibitor | 5 ± 2 | 3 ± 1 |
| TGFB3 | NM_205454 | 47 | TGF beta 3 | 5 ± 0.8 | 5 ± 2.5 |
v-src oncogene was upregulated 265 (+/−45) fold in RSV-SR transformed CEF compared to CEF infected with the non-transforming derivative of RSV-SR td106 mutant. Since there is no v-src gene expressed in the control, non-transformed cells, the over-expression value for the v-src transcript reflects a comparison with the endogenous level of c-src transcript. The value is derived from three independent experiments (n=3) in which chicken genome arrays were screened with cRNA probes derived from independent sets of transformed and infected cells.
“MW” represents molecular weight in kiloDaltons of the unprocessed protein product calculated from the translated cDNA of the predicted gene product with the aid of ExPASy tools.
denotes “similar” in most cases based on the annotation of the sequence provided by Affymetrix. For all the sequences, which we marked here with ‘*’ and therefore termed as “similar” to known mammalian genes, we performed BLASTP analysis and compared them to human proteins. The sequences were from 60 to 80% identical over the main portions of proteins and most likely represent bona fide orthologs. However, a caution is advised, as there are examples of avian-specific genes that share a significant degree of sequence identity with mammalian genes but do not represent their orthologs, e.g., chicken yrk and human yes (Sudol et al., 1993).
N.C. stands for “not confirmed”. It applies to one transcript for which we could not obtain a single size DNA product with the real time PCR, in spite of trying three sets of primers spanning different regions of the sequence.
Gene ontology analysis
In an effort to determine trends in the regulation of classes of genes in response to v-src transformation, lists of Affymetrix Probe Set IDs for 811 genes that are significantly increased or significantly decreased (supplemental Table A) were submitted separately to DAVID 2006 (http://niaid.abcc.ncifcrf.gov/home.jsp). DAVID is a program that facilitates the transition from genome-scale datasets to biological meaning (Dennis et al., 2003). The software identified 41 and 47 terms that are significantly over-represented in a functional annotation chart of the genes with increased or decreased expression, respectively. The most significantly over-represented terms for the genes with increased expression included many related to phosphatase activity, proto-oncogenes, nuclear proteins, and DNA-binding. In contrast, the most significantly over-represented terms for the genes with decreased expression included several related to the cytoskeleton, extracellular matrix, cell adhesion, endoplasmic reticulum, signal peptides and glycoproteins. The Probe Set IDs associated with each of these terms are presented in supplemental Table B. The terms associated with down-regulated genes seem to connote normal cellular growth, attachment, and secretion of regulatory factors, while those associated with up-regulated genes hint at abnormal DNA replication. One stand out feature is the number of phosphatases that are up-regulated in the transformed cells.
Integrating and interpreting the transcriptional fingerprint of v-src in CEFs
Two other approaches were elected to analyze the data globally, focusing on the signaling system. In the first approach, we used the most recent version (2.1) of GenMAPP (Gene MicroArray Pathway Profiler) and MAPPFinder, two software packages from Gladstone Institutes of the University of California at San Francisco (www.genmap.org) to analyze changes in the outcomes of signaling pathways caused by v-src modulated genes. We uploaded 2988 transcripts that were changed by v-src, with p-values no higher than 0.05, into the GenMAPP program and viewed the data on pathway profiles (MAPPFinder) that represent major biological pathways and functional grouping of genes (Doniger et al., 2003). Several signaling pathways, including TGF Beta, MAP kinase, cell cycle, and the kinin-kallikrein component of the complement activation scored well in the GenMAPP analysis in terms of integrating from several to two dozen modulated genes into the signaling schemes. However, the concerted action of changed genes, in terms of activation or repression of the signaling outcomes, was not apparent from this analysis. Therefore, we decided to focus solely on the most robustly changed genes, gathering published data for each one and systematically integrating all of this information into a logical scheme.
Induced genes
After the v-src gene, which we found was overexpressed in the RSV-SR-transformed cells on average 265 fold (compared to cellular src (c-src) gene in RSV-SR-td106 infected CEFs), the urokinase-type plasminogen activator (PLAU) was the second most upregulated gene (196 fold in the array and 42 fold by RT-PCR). Both the PLAU transcript and the protein were previously shown to be elevated in RSV transformed CEFs (Unkeless et al., 1973; Sudol, 1985, Leslie et al., 1990). This enzyme was also implicated in the metastatic potential of tumor cells (Ossowski and Reich, 1980) and the regulation of fibronectin pathway matrix assembly (Monaghan and McKewon-Longo, 2006).
Vasoactive intestinal peptide (VIP) also belongs to the class of genes that are robustly upregulated by v-src (124 fold in the arrays and 53 fold in the RT-PCR) in our screen. As this gene was not known or even expected to be transcriptionally induced in RSV transformed cells, we considered this finding interesting and decided to validate this change further (see below). VIP is a neuromodulator that regulates vasodilation, smooth muscle relaxation and epithelial cell secretion in the intestine. A report on the activation of c-src in cells treated with VIP (Koh, 1992) and a recent study implicating VIP in anti-apoptotic signaling (Sastry et al., 2006) could explain the role of this gene in the enhancement and/or maintenance of oncogenic transformation. The frequent occurrence of c-src activation in human colon cancer (Bolen et al., 1987; Cartwright et al., 1990; Pories et al., 1998) may be functionally linked to the normal expression of intestinal VIP, which may act as a stimulator of src kinase activity (Koh, 1992).
The LOC423497 transcript encodes a protein similar to the human AHNAK (KIAA2019), a high molecular weight (ca. 700 kDa) phosphoprotein that was originally identified as a product of a gene repressed in neuroblastoma cells (Shtivelman et al., 1992; Komuro et al., 2004). One interesting feature of the AHNAK protein is its differential distribution between nuclear versus plasma membrane and cytoplasmic compartments. The nuclear exclusion of AHNAK correlates with cell-to-cell contacts in epithelial cells. The increased expression and the resulting abnormal distribution of the AHNAK in v-src transformed fibroblasts may contribute to the loss of contact inhibition (Haase, 2006).
The induction of transcript LOC423769, which encodes an enzyme similar to human arachidonate 5-lipoxygenase (ALOX5), resonates with an earlier study that documented changes in arachidonic acid metabolism in RSV transformed cells (Barker et al., 1989). The transcript is significantly upregulated; 13 fold in the array experiments and 7 fold in RT-PCR assay. ALOX5 metabolizes arachidonic acid into bioactive lipids, which are generally implicated in inflammation and specifically in colon cancer risk (Goodman et al., 2004; Poole et al., 2006). By promoting inflammation, the ALOX5 gene may contribute to the process of tumor progression.
LOC421692 is v-src induced transcript that is similar to cAMP-specific 3, 5-cyclic phosphodiesterase 7B (PDE7B). PDEs hydrolize the second messengers, cyclic AMP and cyclic GMP, to their corresponding mono-phosphates, affecting a wide spectrum of intercellular events. Studying this particular isoform of PDE in transformed cells and tumors in more detail may uncover a new aspect of signaling by second messengers, which is relevant for oncogenic transformation.
Follistatin (FST) is a small protein that was originally identified by its ability to inhibit FSH (follicle-stimulating hormone) release by binding to activins, the stimulators of FSH secretion. Activins are members of the TGF beta superfamily of ligands. The activins and their antagonist follistatin act as a pleiotropic growth factor system that regulates cell growth and apoptosis (Stove et al., 2004; Krneta et al., 2006). One possible reason for the increase in follistatin levels in RSV transformed cells could be to titrate out activins in order to keep the TGF beta receptor activity low, eliminating its possible growth-inhibiting signals. Decreased levels of TGF beta 3 and the TGF beta-induced gene, TGFBI, both seen in RSV transformed cells, further support this proposal. Moreover, a recent report showed that follistatin is overexpressed in liver tumors and when recombinant follistatin protein was added to normal or pre-neoplastic hepatocytes in primary culture, it stimulated DNA synthesis (Grusch et al., 2006). In the same system, addition of activin repressed the synthesis of DNA. In short, follistatin may promote growth of RSV transformed CEFs.
Dual specificity phosphatases (DUSPs) can target various substrates and dephosphorylate them at serine/threonine and tyrosine residues. DUSPs are known to inactivate MAP kinases by dephosphorylating them at specific tyrosine and threonine sites. Perhaps DUSP5 acts in concert with another gene product induced by v-src: MAP kinase phosphatase 2 (MKP-2). The increase in the expression of MAP kinase phosphatases is in agreement with previous observations (Stofega et al., 1997). In general, cells stably transformed by v-src show only a modest elevation in ERK1/2 activity. However, there is a more marked but transient increase in ERK1/2 activity when a temperature sensitive mutant of v-src is reactivated. Furthermore, v-src-transformed cells are resistant to ERK1/2 activation by TPA or serum. The elevated expression of MKP-2 and DUSP5 could be a part of a negative feedback mechanism which ensures that ERK1/2 activation is only transient (Stofega et al., 1997).
Repressed genes
NOV (nephroblastoma overexpressed) was originally isolated as a gene that was upregulated in Myeloblastosis Associated Virus-induced nephroblastomas in chickens (Joliot et al., 1992; Martinerie et al., 1994). Similar to the PLAU gene, NOV was also previously shown as a transcript that is modulated by v-src in CEFs (Scholz et al., 1996). The NOV (also known as CCN3) gene encodes a secreted protein that is associated with the cellular matrix and promotes cell adhesion. In terms of tumor progression, NOV appears to exhibit suppressive effects in contrast to its related protein, known as CCN (Bleau et al., 2005). Based on the current knowledge of the NOV function in various cells (Lafont et al., 2005; Li et al., 2006; Perbal, 2006), we suggest that the down-regulation of NOV promotes anchorage-independent growth and migration of RSV-SR-transformed CEFs.
Transforming growth factor beta-induced protein of 68 kDa molecular weight (TGFBI) is one of the v-src repressed genes. The TGFBI encodes a protein that is similar to fasciclin-like adhesion protein (Kawamoto et al., 1998). The repression of this gene product most likely contributes to the dramatic reduction in adhesive properties of RSV transformed cells. The substantial down-regulation (5–14 fold) of transforming growth factor beta (TGFB3) itself may provide an additional mechanism that ensures a low level of the TGFBI gene product.
Similar to TGFBI, the repression of “four and a half LIM domain 1” protein (FHL1) should also promote non-anchored growth. A recent study has identified FHL1 gene as a tumor suppressor that acts downstream of src and CAS (focal adhesion adaptor protein Crk-associated substrate) to block anchorage-independent cell growth and migration (Shen et al, 2006). The c-src is known to phosphorylate CAS, which in turn inhibits FHL1 expression and promotes, to a certain degree, cell migration and non-anchored growth. Analogous to the signaling of c-src, the v-src must phosphorylate CAS quite efficiently to block FHL1 expression and to fully enhance non-anchored growth of RSV transformed CEFs. Consistent with this reasoning, FHL1 was found as a gene suppressed in various human tumors (Shen et al., 2006).
The type 2 deiodinase (DIO2) is a seleno-enzyme that catalyzes the conversion of thyroid hormone 4 (thyroxine) to thyroid hormone 3 (triiodothyronine) via deiodination. The repression of this gene in RSV transformed CEFs could lead to the decrease of endogenous triiodothyronine, thereby inhibiting signals of its receptor, cellular erbA. This, in turn, should abrogate inactivation of AP-1 transcription factor as shown previously in chicken cells (Desbois et al., 1991a, b). The active AP-1, together with the upregulated jun (see below), could contribute in unison to signals that enhance cell growth, inflammation and metastasis (Ozanne et al., 2007).
Complement 1q and tumor necrosis factor-related protein 3 (C1QTNF3) is not well characterized in terms of function, and its role in transformation cannot be easily commented on.
In contrast to C1QTNF3, the down-regulation of tissue factor pathway inhibitor gene (TFPI), which encodes a multivalent Kunitz-type serine proteinase inhibitor, could be implicated in the mechanism of transformation. TFPI regulates the extrinsic pathways of blood coagulation. The protein is generally expressed and secreted by vascular cells. Highly malignant human tumors have low or undetectable expression of TFPI compared to benign tumors, implicating the inhibitor in tumor invasion and metastasis (Kondraganti et al., 2006). The activity of TFPI is most likely exerted through plasmin-mediated matrix remodeling. In this light, the acute malignancy of RSV-SR-induced chicken tumors could be explained by the combined action of down-regulated TFPI and up-regulated PLAU.
The model summarizing the concerted action of selected genes that are modulated by v-src is presented in Figure 1.
FIGURE 1. Functional network of robustly induced (red) and repressed (blue) transcripts and their interacting partners (black) in primary chicken cells transformed by Rous Sarcoma Virus.
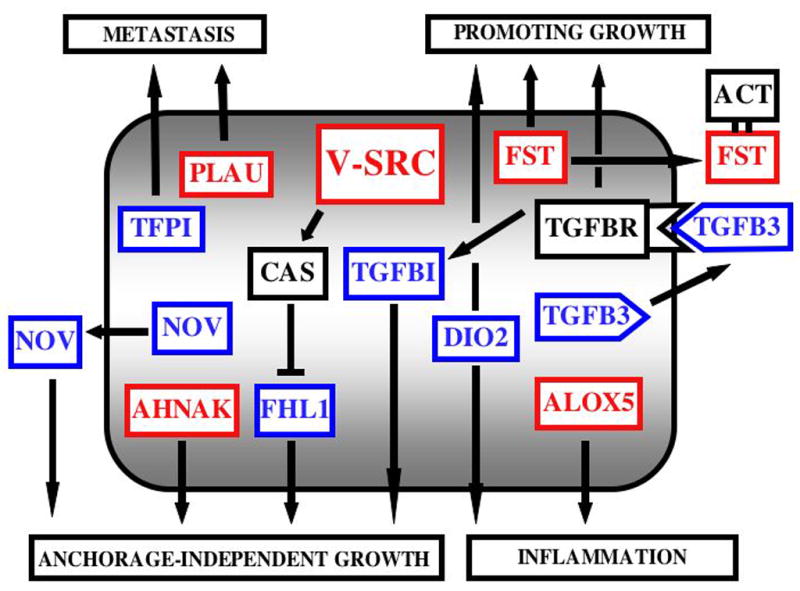
By surveying the literature on the function of each of the 20 gene products that were most dramatically changed in RSV transformed CEFs, we propose a model of their concerted action of promoting cellular transformation and tumor progression. Detailed study of individual genes rather than computer-aided annotation of the genes and their networks was employed to reveal functional outputs of changed transcripts, assuming that changes in transcription corresponded to changes in the protein level. Out of the 20 genes modulated by v-src, 10 could be linked to promoting a cancer phenotype based on their functional characteristics. These include: Follistatin (FST) that contributes to cell growth stimulation by titrating out activin(s) (ACT) and keeping TGF beta receptor (TGFBR) signaling in “low gear”. The low activity of TGFBR is also kept in check by decreased levels of TGF beta 3 (TGFB3). As a consequence, one of the TGF beta-induced genes (TGFBI) that encodes an adhesion protein, is down regulated, contributing to the anchorage-independent growth of transformed cells. Reduced levels of tissue factor pathway inhibitor, (TFPI) together with high levels of urokinase-type plasminogen activator (PLAU), play a role in tumor metastasis. Upregulated arachidonate 5-lipoxygenase (ALOX5) should contribute to tumor development and progression by promoting inflammation. Downregulated type 2 deiodinase (DIO2) would decrease the levels of endogenous tri-iodothyronine and indirectly activate AP-1 transcription factor contributing to signals that enhance cell growth, inflammation and metastasis. The role of four and half LIM domain protein 1 (FHL1) in cell adhesion has been documented and the repression of FHL1, which is regulated by src-phosphorylated CAS, promotes anchorage-independent growth. Tumor suppressive action of nephroblastoma overexpressed gene (NOV) is linked to the control of cell adhesion. The reduced levels of NOV promote anchorage-independent growth. Induction of AHNAK, a gene that encodes a large phosphoprotein, may also contribute to the loss of contact inhibition. See the text for more detailed description of the scheme plus relevant references.
Validating the induction of the VIP transcript at the level of protein
Since up-regulation of the VIP transcript in v-src transformed CEF was substantial (ca. 50 fold), we decided to validate its induction at the level of protein. In parallel, we also performed pilot immuno-stainings of human colon and breast cancer biopsies with src Tyr-416 phospho-specific and VIP antibodies, hoping to see a correlation between the activated src and VIP expression in tumors. Both studies gave us unexpected results. The analysis of VIP protein in RSV-SR transformed cells showed that VIP was also upregulated (9 fold) at the level of protein in transformed cells compared to the control cells that were infected with RSV-SR-td106. However, normal CEFs produced VIP at the level similar to that detected in v-src transformed cells (Figure 2). We concluded that RSV-SR-td106 virus repressed the transcription of the VIP gene. The VIP we detected was around 52 kDa, more than twice the size of the prepro-VIP predicted from the coding sequence of the chicken and human gene. The monoclonal antibody recognized the same size, 52 kDa, protein in the chicken and human brain (data not shown). It is possible that the VIP gene product detected in CEFs and in the brain represents a non-processed and non-secreted form of the hormone or may encode a new VIP paralog.
FIGURE 2. Immunoblot analysis of VIP in normal and RSV-SR transformed CEFs.
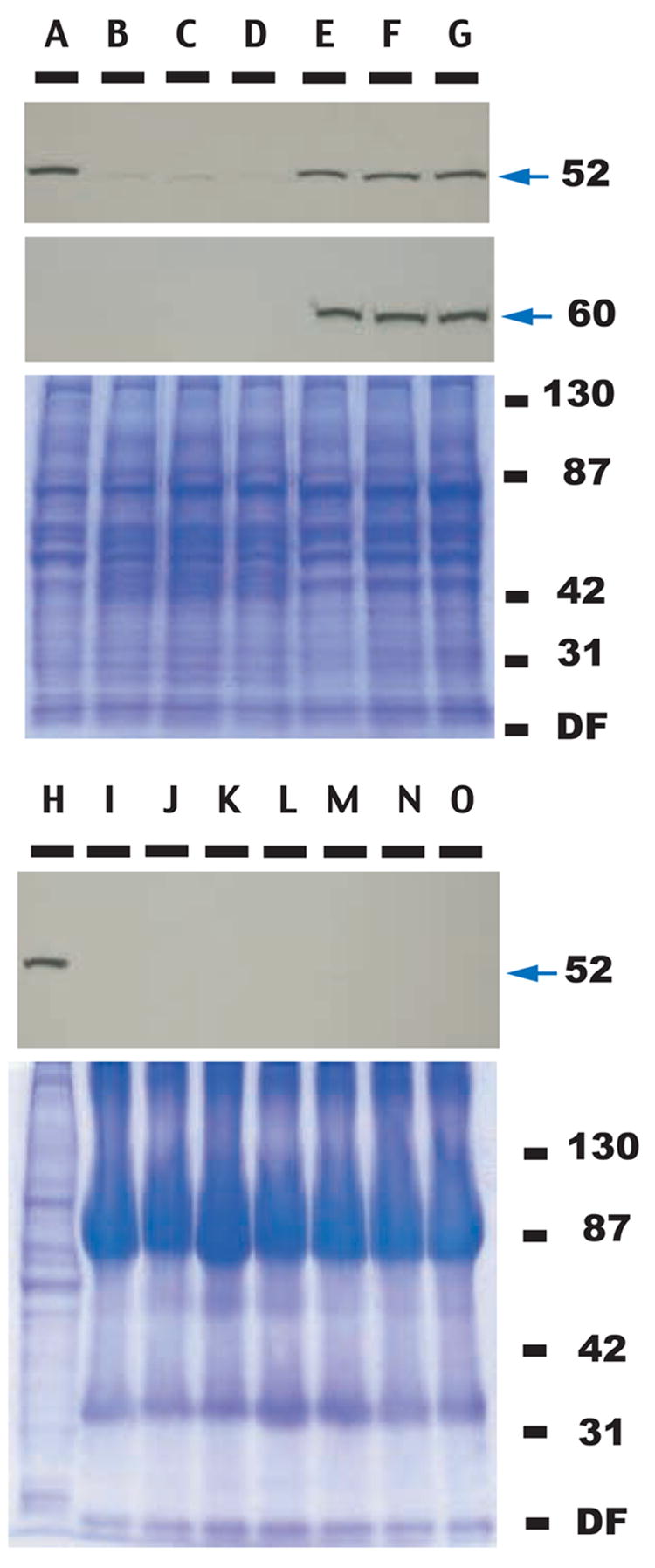
Upper three-partite panel, top panel (lanes A–G): Western blot analysis of cell lysates from CEFs (lane A), CEFs infected with RSV-SR td106 mutant (lanes B, C, D), and CEFs transformed by RSV-SR (lanes E, F, G). The blot was probed with a monoclonal antibody raised against human VIP. Lysates from cells derived from three independent experiments (infection with RSV-SR td106 mutant and transformation with RSV-SR) were used. Arrow points to VIP of 52 kDa molecular weight. Middle panel, the same Western blot was probed with src phospho-Tyr 416-specific antibody. Arrow points to c-src of 60 kDa molecular weight. Lower panel, Coomassie blue staining of the parallel SDS-polyacrylamide gel analyzed in the upper panels.
Lower two panels, top panel (lanes H–O): Western blot analysis of conditioned media from normal and RSV-SR transformed CEFs; lane H, lysate from normal, uninfected CEFs that serves as a positive control for the presence of VIP; lanes I–K, supernatant from CEFs infected with RSV-SR td 106 mutant; and lanes M–O, supernatant from CEFs transformed with RSV-SR. Arrow indicates VIP migrating at the apparent 52 kDa molecular weight. Lower panel, Coomassie blue staining of the parallel SDS-polyacrylamide gel analyzed in the upper panel.
Staining of 12 colon carcinomas and 13 breast carcinoma biopsies identified tumor tissues with clear immunoreactivity for src Tyr-416 phospho-specific antibodies (8 in colon adenocarcinoma and 9 in breast carcinoma), yet none of the 25 biopsies showed staining for VIP (Jianhui Shi, Fan Lin and Conrad Schuerch, unpublished data). As expected, the VIP antibody showed staining in ganglion cells residing in normal sections of colon.
General concerns and concluding remarks
The aim of our study was to survey transcriptional changes affected by RSV and its transforming gene, v-src, in primary cultures of CEFs. Apart from the basic insight into the process of transformation, we wanted to identify robustly changed transcripts that could serve as cancer biomarkers. Several genes that were not previously known as transcripts changed in transformed cells or sarcomas were identified. Among them, is FST, a hormone-like substance whose protein level could be monitored in human blood.
Two other studies reported analysis of v-src-affected genes (Malek et al., 2001; Paz et al., 2004). However, both analyses used a xenogeneic system of chicken v-src that was overexpressed in established rodent cell lines, 3Y1 and NIH3T3 lines. Both studies used cells selected with the aid of antibiotic resistance genes for src expression, following transfection. In addition, the arrays used in the studies contained a limited number of genes (8,900 human genes in Malek et al. report; and 12,000 mouse sequences in which 6,000 were non-annotated ESTs in Paz et al. report). Moreover, one of the two studies documented a concordance between transcripts that were present in src transformed 3Y1 cells and human tumors, but did not show direct changes between v-src transformed and non-transformed 3Y1 cells (Malek et al., 2004). Because of these factors, it is difficult to compare the transcriptional fingerprint of v-src transformation of CEFs that we generated to that reported previously in rodent cells, especially in the selected set of 20 robustly modulated genes. With the exception of one robustly down-regulated gene that encodes LIM protein 1 (FHL1), which was also identified by Paz and colleagues (Paz et al., 2004), the remaining 19 genes we reported here (Table 1) were not scored in previous array screens.
We also identified the less robust changes in v-src affected transcripts (Table A, Supplementary Material) and consider them important for the understanding of transformation fingerprint. Small changes (two to three fold modulation) in the transcription of genes that encode regulatory proteins or transcription factors per se could result in unregulated signaling and contribute to the transformation process (Vogt, 2001). The 3-fold upregulation of jun and myc transcripts was recorded in the array screen. Small changes in the level of transcripts encoding proteins that act in the same pathway could produce synergistic responses. The best example in our data is represented by vascular endothelial growth factor (VEGF) and ribosomal protein S6 kinase. These two genes are up-regulated 3 fold each and their protein products participate in the mTOR pathway (Klos et al., 2006). In RSV-SR transformed CEFs, the level of VEGF gene product is additionally increased by ribosomal protein S6 kinase, which enhances translation of VEGF RNA. The resulting synergy between the two up-regulated gene products may contribute significantly to angiogenesis and metastasis of v-src induced tumors. Similarly, we confirmed the upregulation of Eps8 transcript, whose protein serves as a substrate of v-src kinase (Gallo et al., 1997). The concerted action of v-src and Eps8 should contribute to signals stimulating cell growth.
The transcriptional fingerprint of RSV transformation reported here represents a static and limited view of the transformation process. We have isolated total RNAs from cells that were fully transformed or infected. The cells were harvested soon after they reached complete transformation, as judged by visual inspection. To generate a dynamic picture of transcriptional changes caused by v-src, it would be necessary to use temperature-sensitive mutants of RSV. It is likely that certain genes are only modulated at the initial and narrow window of time just when the threshold of transformation by v-src is reached (Jakobovits et al., 1984; Jahner and Hunter, 1991). It would also be important to look at genes modulated in RSV transformed cells that were propagated for longer periods of time, allowing adaptation and the epigenetic changes that must occur in spontaneous cancer. Such experiments would allow us to differentiate between genes that are primary targets of v-src-induced nuclear signaling pathways and genes that are modulated by the subsequent wave of “secondary” events. Finally, to obtain a more complete picture of transformation, one should connect the repertoire of changed genes with the signaling network of src as a protein-tyrosine kinase (Blume-Jensen and Hunter, 2001; Feng et al., 2006).
In our survey, we confirmed two out of nine v-src-induced genes that were identified in rat fibroblasts transformed by RSV LA23 temperature sensitive mutant (Chen et al., 1977: Jahner and Hunter, 1991). These two genes were rhoB and ornithine decarboxylase. In our assay, both transcripts were elevated only two fold in transformed cells and therefore were excluded from the ‘stringent’ list of 811 genes. An interesting observation should be noted here. In rat fibroblasts, v-src induced a gene, which encoded tissue factor. We did not observe the induction of the tissue factor transcript in CEFs although its probe was present and detected in the array. We did, however, record the repression of tissue factor pathway inhibitor (see Table 1). This single observation could be an example of v-src “resourcefulness” in transforming fibroblasts of different species. Src does this by targeting different regulators of the same pathway to achieve the same final “read-out”. Host-specific differences in signaling by src in chicken versus rodent cells were well documented in the past, and our study further supports this phenomenon (e.g., Hirai and Varmus, 1990).
Interleukin 8 (9E3) and gravin are two genes modulated in RSV transformed cells (Sugano et al., 1987; Bedard et al., 1987; Frankfort and Gelman, 1995). These genes were also scored for in our arrays and their transcripts changed as reported previously, yet the fold changes that we observed were below the two and a half fold cut off.
There are three concerns which need to be addressed in future studies. First, it is important to stress that the mRNA levels do not always correlate with the protein levels (e.g. Tian et al., 2004). Therefore, one needs to show that changes in the level of transcript, which we report here, correspond to changes in the level of protein. Second, the primary culture of chicken embryo fibroblasts is known to also contain small numbers of additional cell types that may respond differently to the RSV infection. The presence of small numbers of myoblasts, neurons, epithelial and perhaps stem cells that ‘contaminate‘ the dominant population of fibroblasts may be responsible for detection of transcripts which seem uncommon for fibroblasts (e.g. nestin). The use of immortalized CEF cell lines such as DF-1 and SC-1 may not provide the best solution to the problem as these cells have unique, immortalization- and transformation-related changes reflected in the transcription of mitochondria-encoded genes (DF-1) or alterations in the p53 and Rb pathways (SC-1) (Kim et al., 2001; Christman et al., 2005). Third, our control virus containing v-src deletion (RSV-SR-td106) showed strong activity in repressing the VIP gene in CEFs. Other v-src-regulated transcripts could also have been affected by the non-transforming virus; therefore, one should use the primary, non-infected CEFs to control for such a possibility.
In sum, we have identified and confirmed a set of twelve new genes that are highly induced (seven) or repressed (five) in Rous Sarcoma Virus transformed primary chicken embryo cells, compared to cells infected with the control virus with deleted src gene. The quantitative changes and the function of the modulated genes should help us in better understanding the molecular mechanism of oncogenic transformation, which for v-src oncogene is characterized by dramatic neoplastic changes of primary cells and high malignancy of resulting tumors. Despite the significant phylogenetic distance of 310 million years between birds and mammals (Burt, 2005), and examples of species-specific differences in signaling between certain orthologs (Van Dyke and Jacks, 2002; Vogelstein and Kinzler, 2006), the human homologues of at least some of the v-src modulated chicken genes could represent biomarkers of human cancers (Russello and Shore, 2004; Homsi et al., 2007).
Materials and methods
Cells, Viruses and Preparation of RNA
Chicken embryo fibroblasts (CEFs) were prepared and maintained as previously described (Hanafusa 1969). Three sets of CEFs were infected with high-titer stocks of two viruses. Half of the plates were infected with the transforming, oncogenic virus - Schmidt-Ruppin strain of RSV subgroup A (RSV-SR). The other half were infected with the non-transforming derivative of RSV-SR named td106, in which the src gene was deleted (Wang et al., 1984). Both sets of cells were propagated in parallel under standard conditions. When RSV-SR CEFs showed complete morphological transformation (Hanafusa, 1969), the total RNA from the transformed cells and from the control cells infected with td106 RSV-SR was isolated using Qiagen Rneasy Minikit. The RNA was isolated from sub-confluent cultures to ensure that differences in the growth rate between normal and transformed cells were not affected by contact inhibition of normal, confluent CEFs. Yield and quality of RNA were determined through measurement of the absorbance at 260 nm and 280 nm. All of the RNAs used in the experiments had a 260/280 ratio of 1.9 or higher. These preparations of total RNA served as the template for cDNA synthesis and subsequently for the generation of biotinylated cRNA probes that were used directly in the hybridization to Chicken GeneChipR Genome Arrays [Lot # 4009764 and 4017168] (Affymetrix, Inc., Santa Clara, CA).
Microarray Analysis
For each chip, 5μg of RNA was used to synthesize cDNA with an oligo-dT primer attached to a sequence of the T7 promoter region. An in-vitro transcription reaction was used to generate biotin-labeled cRNA from the cDNA. The cRNA was fragmented and an agarose gel was run to verify the sizes of the RNA fragments. A hybridization cocktail was then prepared using 20 μg of the fragmented cRNA. The cocktail was hybridized to a Chicken GeneChipR Genome array at 45°C for 18 hr. Next, the cocktail was removed and the arrays were washed and incubated with R-Streptavidin-Phycoerithrin (Molecular Probes, Eugene, OR) following the protocol supplied by the manufacturer. The signal was amplified using a biotinylated antibody raised against streptavidin (Vector Laboratories, Burlingham, CA). The arrays were then washed, stained, and scanned following standard Affymetrix protocols. Each total RNA preparation and its derived cRNA was used to probe one array, giving rise to three experimental values for each chicken transcript that hybridized with the probe. The results were analyzed with the Affymetrix GeneChip Operating Software 2.1 and Spotfire Decision Site 8.1.
Data Analysis
Affymetrix GeneChip Operating Software 2.1 (GCOS) standard adjustments for artifacts, noise and background were used to calculate a detection p-value, detection call and signal for each gene on each array. The entire data set was imported into Microsoft Excel where it was sorted by the detection call. All gene probes scoring a present or marginal on two or more of the arrays in each group were selected for further analysis. This data was imported into Spotfire Decision Site 8.1 where the first analysis was completed. The data set was normalized by scaling each experiment by mean, followed by z-score calculation of the gene probes. The resulting data was then analyzed using a one-way analysis of variance (ANOVA) test to determine which genes were significantly changed. A p-value of ≤ 0.05 was considered significant. The Spotfire Software was then used to cluster the experiments (columns) and gene probes (rows) into a heat map using the unweighted average and Euclidean distance methods. The data was also analyzed using the Affymetrix GCOS software. This consisted of taking the original data from each of the three experiments in the entire data set and comparing them separately. The natural log fold change for each experiment was calculated using the control as the baseline. This data was imported to Microsoft Excel, where the log change was converted to linear change. The final data set consisted of those genes, which had at least a 2.5 linear fold change from the GCOS analysis and a p-value of less than 0.05 from the Spotfire analysis.
Quantitative Real-Time PCR
New total RNAs were prepared from two sets of SR-A-RSV transformed CEFs and two sets of CEFs infected with SR-A-RSV td106. Both sets of RNA from each condition were pooled to make cDNA. The cDNA was synthesized using 2 μg of RNA, 8μl of 5x First Strand Buffer (Invitrogen, Carlsbad, CA), 2μl of 0.1M DTT, 1μl of 50μM Random Hexamers, 0.8μl of 25μM dNTPs, 1μl of Superscript II RT (Invitrogen, Carlsbad, CA), and PCR grade water to bring the final volume of each reaction to 40μl. PCR conditions were 22 °C for 10 minutes, 40 °C for 40 minutes, 99 °C for 5 minutes and 10°C hold. Quantitative Real-Time PCR (RT-PCR) was performed using Quantitect SYBR Green PCR kit (Qiagen, Valencia, CA) as per manufacturer-suggested protocol. A standard curve of serial dilutions (1:1 – 1:100000 of cDNA) and a no template control were included for each primer pair. RT-PCR analysis was completed using the second derivative maximum method of Roche Molecular Biochemicals Light-cycler Software version 3.5 (Roche, Pleasanton, CA). Using this method, a relative concentration for each sample was generated based on the standard curve of the target of interest. This value was then normalized to the corresponding relative concentration of GAPDH. The resulting normalized data was further used to determine the change in expression of each target by taking the ratio of the experimental value to the control value. Sets of 22-mer-long forward [F] and reverse [R] primers for the chicken GAPDH and the 20 individual transcripts were designed to amplify DNA fragments below 200 bp long. Integrated DNA Technologies Company synthesized and provided the oligo-nucleotides. The primers used in the RT-PCR assay were as follows:
HOUSEKEEPER - GAPDH1-F – AATGCATCGTGCACCACCAACT;
HOUSEKEEPER - GAPDH1-R – TCCCATTCAGCTCAGGGATGACTT;
PLAU-F – TGCAACTTGGACTGGGCAAACA;
PLAU-R – TGCAGGGCGTTTCTTGAATGGA;
VIP_F- TGGGAAACAGACTGCCCTTTGA;
VIP_R- AGCATCAGAGTGGCGTTTGACA;
LOC423497_F-ACATTGCCGTGGAAGCACCAAA;
LOC423497_R-AATCTCACCCGTTGGGAAGTGAGT LOC421581_F-ATCCTGATGTACGCCTGGTTCA;
LOC421581_R-ATTAGGAGAGCAGCAGGCAACT;
LOC423769_F- TTTGGCCAGTATGACTGGTGCT;
LOC423769_R- AACGCTGTTGGGAATGCGGTTT;
LOC421692_F-TGTCTGCATGGCAACAGCATCA;
LOC421692_R-TTGATGAACAGCTGGGTGAGGGAT;
FST_F- TGTGTTTGTGCTCCGGATTGCT;
FST_R- TTTGATCCACCACACACGTGGA;
LOC423890_F- TTGAAGGCTCGTGTTTCGCCTT;
LOC423890_R- TTCAGCAGGATGAAGAAGCCGT;
NES_F- AGCACACCCATTTCCCTTTGGT;
NES_R- AGTGGGAGGGCGATGCTTTATT;
MKP2_F- AAGAAGCGCGTCAAGCTGGAAA;
MKP2_R- AATGGCAGTGTCCGGCACTATT;
NPC2_F- AAGCAGCTGGATAAGAGGGCAT;
NPC2_R-TTGTTGTTCTCCTCCAGTGCCT;
NOV_F-TGCACCAGTGTGCAGACTTACA;
NOV_R-TGAGGACAGCGGAACTCAACTT;
TGFBI_F-GAAGCTTTCCGAGCCATGCCAC;
TGFBI_R -TGGACTTGAGCCGCACAAGAGC;
LOC418180_F-ACGCCGAATGCATCTACACGAA;
LOC418180_R-AGCCAACTTCTCTCATGTGGCA;
LOC428711_F-AAGCAATTGGGTGGGAAGCGTT;
LOC428711_R-TCTTGCAGCCAGCACACTTCTT;
DIO2_F- AAGGGCAACGATGGCAGCAATA;
DIO2_R-TGCACAATGCACACTCGCTCAA;
C1QTNF3_F- ATCAGTTGGCATCTGCTGGCTT;
C1QTNF3_R- TTCTGGAACCTTTGTGGCCAGT;
CPEB1_F-TGCAGCACAAACTAGCCCACAA;
CPEB1_R-ACTGCAGCCAAAGCAGCTTGAT;
TFPI_F-ATGCCATTGCCCTTGGTCTTGA;
TFPI_R-AGTCATCAGTTGCATGCCCAGT;
TGFB3_F-AGAACTGTAATCCAGGAGGCCTGT;
TGFB3_R-TGGGAGCCTTGCAGTTTGCATT.
Immunoblot analysis
Lysates or clarified culture media of uninfected CEFs, of CEFs infected with RSV-SR mutant td-106, and of CEFs transformed by RSV-SR were fractionated on 4–20% pre-cast SDS-polyacrylamide electrophoresis iGels (NG11–420 Life Therapeutics, Australia). Tissues and cells were homogenized in RIPA lysis buffer containing a cocktail of mammalian protease inhibitors. Lysates of adult, normal chicken and human tissues (brain, colon and intestine) were obtained from Zyagen Laboratories (San Diego, CA). Lysate samples: were 25ul/well (CEFs - 40 ug, chicken and human tissues – 100 ug); media samples were: 5ul/well (200 ug). As molecular weight markers we used Kaleidoscope Prestained Standard (161-0324 BioRad, Hercules, CA).
Transfer of resolved proteins to nitrocellulose membrane was monitored by Pounceau red staining. Normalization of proteins was completed on parallel gels and visualized by staining with Coomassie Brilliant Blue R250 solution (Bio-Rad Laboratories, Inc., Hercules, CA) following a standard protocol. After transfer, lysate proteins on nitrocellulose membranes were incubated with anti-VIP monoclonal antibody (SC-25347, Santa Cruz) at 1:200 dilution. The secondary antibody was peroxidase-labeled anti-mouse (NIF825, Amersham) at 1: 5000. After transfer, media proteins on nitrocellulose membranes were incubated with anti-VIP (1:100), with peroxidase-labeled anti-mouse (1:3500). Both blots were then stripped at 65.0°C for 30 minutes, washed 2 times for 10 minutes with TBST containing 0.05% Tween 20 and then blocked one hour at RT in 5% milk. The blots were then incubated with anti-phospho-Src antibody (Tyr 416)(2101S, Cell Signaling) at 1:625 dilution followed by the peroxidase-labeled anti-rabbit secondary antibody (1:10,000). ECL cheminoluminescence kit from Amersham (RPN2108) was used for detection of proteins on Western blots.
To quantify the relative differences in the expression of proteins, auto-radiographs of Western blots were scanned using Imaging System unit from Alpha Innotech. Spot Densitometry Quantitation Tool program was used.
Supplementary Material
To identify sets of genes on the basis of similarities in the pattern with which their expression changed in RSV transformed cells, we applied two-dimensional hierarchical clustering. We considered all the genes with p-value of less than 0.05 and the linear fold change more that 2.5. The Spotfire Software was then used to cluster the experiments (columns) and gene probes (rows) into a heat map using the un-weighted average and Euclidean distance methods.
List of genes and open reading frames that are induced (I) or repressed (D, after decrease) for equal or more than 2.5 fold (linear, in either direction) and with the p-value less than 0.05 in RSV-SR transformed CEFs compared to RSV-SR td106 mutant infected CEFs.
Summary of gene ontology terms associated with transcripts significantly modulated by v-src.
Acknowledgments
We would like to thank Gary Bader, Angelika Barnekow, Glenn Gerhard, Tony Hunter, Steve G. Martin, Hisataka Sabe, Jacques Samarut, Wolf-Dieter Schleuning, Momin M. Shareef and Carter Van Waes for constructive comments on the first version of the manuscript. Fan Lin, Jianhui Shi and Conrad Schuerch are acknowledged for sharing unpublished data. This study was supported by a grant from Pennsylvania Department of Health (DJC) and NIH grants CA29339 (LHW), DK62345 (MS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barker K, Aderem A, Hanafusa H. Modulation of arachidonic acid metabolism by Rous sarcoma virus. J Virol. 1989;63:2929–2935. doi: 10.1128/jvi.63.7.2929-2935.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard PA, Alcorta D, Simmons DL, Luk KC, Erikson RL. Constitutive expression of a gene encoding a polypeptide homologous to biologically active human platelet protein in Rous sarcoma virus-transformed fibroblasts. Proc Natl Acad Sci U S A. 1987;84:6715–6719. doi: 10.1073/pnas.84.19.6715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau AM, Planque N, Perbal B. CCN proteins and cancer: two to tango. Front Biosci. 2005;10:998–1009. doi: 10.2741/1594. [DOI] [PubMed] [Google Scholar]
- Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- Bolen JB, Veillette A, Schwartz AM, DeSeau V, Rosen N. Activation of pp60c-src protein kinase activity in human colon carcinoma. Proc Natl Acad Sci U S A. 1987;84:2251–2255. doi: 10.1073/pnas.84.8.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brugge JS, Erikson RL. Identification of a transformation-specific antigen induced by an avian sarcoma virus. Nature. 1977;269:346–348. doi: 10.1038/269346a0. [DOI] [PubMed] [Google Scholar]
- Burt DW. Chicken genome: current status and future opportunities. Genome Res. 2005;15:1692–1698. doi: 10.1101/gr.4141805. [DOI] [PubMed] [Google Scholar]
- Cartwright CA, Meisler AI, Eckhart W. Activation of the pp60c-src protein kinase is an early event in colonic carcinogenesis. Proc Natl Acad Sci U S A. 1990;87:558–562. doi: 10.1073/pnas.87.2.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Hayman MJ, Vogt PK. Properties of mammalian cells transformed by temperature-sensitive mutants of avian sarcoma virus. Cell. 1977;11:513–521. doi: 10.1016/0092-8674(77)90069-1. [DOI] [PubMed] [Google Scholar]
- Christman SA, Kong BW, Landry MM, Kim H, Foster DN. Modulation of p53 expression and its role in the conversion to a fully immortalized chicken embryo fibroblast line. FEBS Lett. 2005;579:6705–6715. doi: 10.1016/j.febslet.2005.10.066. [DOI] [PubMed] [Google Scholar]
- Darnell JE., Jr Transcription factors as targets for cancer therapy. Nat Rev Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- Dehbi M, Bedard PA. Regulation of gene expression in oncogenically transformed cells. Biochem Cell Biol. 1992;70:980–997. doi: 10.1139/o92-142. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- Desbois C, Aubert D, Legrand C, Pain B, Samarut J. A novel mechanism of action for v-ErbA: abrogation of the inactivation of transcription factor AP-1 by retinoic acid and thyroid hormone receptors. Cell. 1991;67:731–740. doi: 10.1016/0092-8674(91)90068-a. [DOI] [PubMed] [Google Scholar]
- Desbois C, Pain B, Guilhot C, Benchaibi M, Ffrench M, Ghysdael J, Madjar JJ, Samarut J. v-erbA oncogene abrogates growth inhibition of chicken embryo fibroblasts induced by retinoic acid. Oncogene. 1991;6:2129–2135. [PubMed] [Google Scholar]
- Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagan JB, Sobel ME, Yamada KM, de Crombrugghe B, Pastan I. Effects of transformation on fibronectin gene expression using cloned fibronectin cDNA. J Biol Chem. 1981;256:520–525. [PubMed] [Google Scholar]
- Feng Q, Baird D, Peng X, Wang J, Ly T, Guan JL, Cerione RA. Cool-1 functions as an essential regulatory node for EGF receptor- and Src-mediated cell growth. Nat Cell Biol. 2006;8:945–956. doi: 10.1038/ncb1453. [DOI] [PubMed] [Google Scholar]
- Frame MC. Src in cancer: deregulation and consequences for cell behaviour. Biochim Biophys Acta. 2002;1602:114–130. doi: 10.1016/s0304-419x(02)00040-9. [DOI] [PubMed] [Google Scholar]
- Frankfort BJ, Gelman IH. Identification of novel cellular genes transcriptionally suppressed by v-src. Biochem Biophys Res Commun. 1995;206:916–926. doi: 10.1006/bbrc.1995.1130. [DOI] [PubMed] [Google Scholar]
- Gallo R, Provenzano C, Carbone R, Di Fiore PP, Castellani L, Falcone G, Alema S. Regulation of the tyrosine kinase substrate Eps8 expression by growth factors, v-Src and terminal differentiation. Oncogene. 1997;15:1929–1936. doi: 10.1038/sj.onc.1201344. [DOI] [PubMed] [Google Scholar]
- Goodman JE, Bowman ED, Chanock SJ, Alberg AJ, Harris CC. Arachidonate lipoxygenase (ALOX) and cyclooxygenase (COX) polymorphisms and colon cancer risk. Carcinogenesis. 2004;25:2467–2472. doi: 10.1093/carcin/bgh260. [DOI] [PubMed] [Google Scholar]
- Grusch M, Drucker C, Peter-Vorosmarty B, Erlach N, Lackner A, Losert A, Macheiner D, Schneider WJ, Hermann M, Groome NP, et al. Deregulation of the activin/follistatin system in hepatocarcinogenesis. J Hepatol. 2006 doi: 10.1016/j.jhep.2006.06.014. [DOI] [PubMed] [Google Scholar]
- Haase H. Ahnak, a new player in beta-adrenergic regulation of the cardiac L-type Ca(2+) channel. Cardiovasc Res. 2007;73:19–25. doi: 10.1016/j.cardiores.2006.09.001. [DOI] [PubMed] [Google Scholar]
- Hanafusa H. Rapid transformation of cells by Rous sarcoma virus. Proc Natl Acad Sci U S A. 1969;63:318–325. doi: 10.1073/pnas.63.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendricks M, Weintraub H. Tropomyosin is decreased in transformed cells. Proc Natl Acad Sci U S A. 1981;78:5633–5637. doi: 10.1073/pnas.78.9.5633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herault Y, Chatelain G, Brun G, Michel D. V-src-induced-transcription of the avian clusterin gene. Nucleic Acids Res. 1992;20:6377–6383. doi: 10.1093/nar/20.23.6377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillier LW, Miller W, Birney E, Warren W, Hardison RC, Ponting CP, Bork P, Burt DW, Groenen MA, Delany ME, et al. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Hirai H, Varmus HE. Mutations in src homology regions 2 and 3 of activated chicken c-src that result in preferential transformation of mouse or chicken cells. Proc Natl Acad Sci U S A. 1990;87:8592–8596. doi: 10.1073/pnas.87.21.8592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsi J, Cubitt C, Daud A. The Src signaling pathway: a potential target in melanoma and other malignancies. Expert Opin Ther Targets. 2007;11:91–100. doi: 10.1517/14728222.11.1.91. [DOI] [PubMed] [Google Scholar]
- Howard BH, Adams SL, Sobel ME, Pastan I, de Crombrugghe B. Decreased levels of collagen mRNA in Rous sarcoma virus-transformed chick embryo fibroblasts. J Biol Chem. 1978;253:5869–5874. [PubMed] [Google Scholar]
- Jahner D, Hunter T. The stimulation of quiescent rat fibroblasts by v-src and v-fps oncogenic protein-tyrosine kinases leads to the induction of a subset of immediate early genes. Oncogene. 1991;6:1259–1268. [PubMed] [Google Scholar]
- Jakobovits EB, Majors JE, Varmus HE. Hormonal regulation of the Rous sarcoma virus src gene via a heterologous promoter defines a threshold dose for cellular transformation. Cell. 1984;38:757–765. doi: 10.1016/0092-8674(84)90271-x. [DOI] [PubMed] [Google Scholar]
- Joliot V, Martinerie C, Dambrine G, Plassiart G, Brisac M, Crochet J, Perbal B. Proviral rearrangements and overexpression of a new cellular gene (nov) in myeloblastosis-associated virus type 1-induced nephroblastomas. Mol Cell Biol. 1992;12:10–21. doi: 10.1128/mcb.12.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jove R, Hanafusa H. Cell transformation by the viral src oncogene. Annu Rev Cell Biol. 1987;3:31–56. doi: 10.1146/annurev.cb.03.110187.000335. [DOI] [PubMed] [Google Scholar]
- Kawamoto T, Noshiro M, Shen M, Nakamasu K, Hashimoto K, Kawashima-Ohya Y, Gotoh O, Kato Y. Structural and phylogenetic analyses of RGD-CAP/beta ig-h3, a fasciclin-like adhesion protein expressed in chick chondrocytes. Biochim Biophys Acta. 1998;1395:288–292. doi: 10.1016/s0167-4781(97)00172-3. [DOI] [PubMed] [Google Scholar]
- Kim H, You S, Kim IJ, Farris J, Foster LK, Foster DN. Increased mitochondrial-encoded gene transcription in immortal DF-1 cells. Exp Cell Res. 2001;265:339–347. doi: 10.1006/excr.2001.5207. [DOI] [PubMed] [Google Scholar]
- Klos KS, Wyszomierski SL, Sun M, Tan M, Zhou X, Li P, Yang W, Yin G, Hittelman WN, Yu D. ErbB2 increases vascular endothelial growth factor protein synthesis via activation of mammalian target of rapamycin/p70S6K leading to increased angiogenesis and spontaneous metastasis of human breast cancer cells. Cancer Res. 2006;66:2028–2037. doi: 10.1158/0008-5472.CAN-04-4559. [DOI] [PubMed] [Google Scholar]
- Koh SW. The pp60c-src in retinal pigment epithelium and its modulation by vasoactive intestinal peptide. Cell Biol Int Rep. 1992;16:1003–1014. doi: 10.1016/s0309-1651(06)80053-6. [DOI] [PubMed] [Google Scholar]
- Komuro A, Masuda Y, Kobayashi K, Babbitt R, Gunel M, Flavell RA, Marchesi VT. The AHNAKs are a class of giant propeller-like proteins that associate with calcium channel proteins of cardiomyocytes and other cells. Proc Natl Acad Sci U S A. 2004;101:4053–4058. doi: 10.1073/pnas.0308619101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondraganti S, Gondi CS, Gujrati M, McCutcheon I, Dinh DH, Rao JS, Olivero WC. Restoration of tissue factor pathway inhibitor inhibits invasion and tumor growth in vitro and in vivo in a malignant meningioma cell line. Int J Oncol. 2006;29:25–32. [PMC free article] [PubMed] [Google Scholar]
- Krneta J, Kroll J, Alves F, Prahst C, Sananbenesi F, Dullin C, Kimmina S, Phillips DJ, Augustin HG. Dissociation of angiogenesis and tumorigenesis in follistatin- and activin-expressing tumors. Cancer Res. 2006;66:5686–5695. doi: 10.1158/0008-5472.CAN-05-3821. [DOI] [PubMed] [Google Scholar]
- Lafont J, Thibout H, Dubois C, Laurent M, Martinerie C. NOV/CCN3 induces adhesion of muscle skeletal cells and cooperates with FGF2 and IGF-1 to promote proliferation and survival. Cell Commun Adhes. 2005;12:41–57. doi: 10.1080/15419060500383069. [DOI] [PubMed] [Google Scholar]
- Leslie ND, Kessler CA, Bell SM, Degen JL. The chicken urokinase-type plasminogen activator gene. J Biol Chem. 1990;265:1339–1344. [PubMed] [Google Scholar]
- Li CL, Coullin P, Bernheim A, Joliot V, Auffray C, Zoroob R, Perbal B. Integration of Myeloblastosis Associated Virus proviral sequences occurs in the vicinity of genes encoding signaling proteins and regulators of cell proliferation. Cell Commun Signal. 2006;4:1. doi: 10.1186/1478-811X-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek RL, Irby RB, Guo QM, Lee K, Wong S, He M, Tsai J, Frank B, Liu ET, Quackenbush J, et al. Identification of Src transformation fingerprint in human colon cancer. Oncogene. 2002;21:7256–7265. doi: 10.1038/sj.onc.1205900. [DOI] [PubMed] [Google Scholar]
- Martin GS. The road to Src. Oncogene. 2004;23:7910–7917. doi: 10.1038/sj.onc.1208077. [DOI] [PubMed] [Google Scholar]
- Martinerie C, Huff V, Joubert I, Badzioch M, Saunders G, Strong L, Perbal B. Structural analysis of the human nov proto-oncogene and expression in Wilms tumor. Oncogene. 1994;9:2729–2732. [PubMed] [Google Scholar]
- Monaghan-Benson E, McKeown-Longo PJ. Urokinase-type plasminogen activator receptor regulates a novel pathway of fibronectin matrix assembly requiring Src-dependent transactivation of epidermal growth factor receptor. J Biol Chem. 2006;281:9450–9459. doi: 10.1074/jbc.M501901200. [DOI] [PubMed] [Google Scholar]
- Ossowski L, Reich E. Experimental model for quantitative study of metastasis. Cancer Res. 1980;40:2300–2309. [PubMed] [Google Scholar]
- Ozanne BW, Spence HJ, McGarry LC, Hennigan RF. Transcription factors control invasion: AP-1 the first among equals. Oncogene. 2007;26:1–10. doi: 10.1038/sj.onc.1209759. [DOI] [PubMed] [Google Scholar]
- Paz K, Socci ND, van Nimwegen E, Viale A, Darnell JE. Transformation fingerprint: induced STAT3-C, v-Src and Ha-Ras cause small initial changes but similar established profiles in mRNA. Oncogene. 2004;23:8455–8463. doi: 10.1038/sj.onc.1207803. [DOI] [PubMed] [Google Scholar]
- Perbal B. NOV story: the way to CCN3. Cell Commun Signal. 2006;4:3. doi: 10.1186/1478-811X-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole EM, Bigler J, Whitton J, Sibert JG, Potter JD, Ulrich CM. Prostacyclin synthase and arachidonate 5-lipoxygenase polymorphisms and risk of colorectal polyps. Cancer Epidemiol Biomarkers Prev. 2006;15:502–508. doi: 10.1158/1055-9965.EPI-05-0804. [DOI] [PubMed] [Google Scholar]
- Pories SE, Hess DT, Swenson K, Lotz M, Moussa R, Steele G, Jr, Shibata D, Rieger-Christ KM, Summerhayes C. Overexpression of pp60c-src elicits invasive behavior in rat colon epithelial cells. Gastroenterology. 1998;114:1287–1295. doi: 10.1016/s0016-5085(98)70435-4. [DOI] [PubMed] [Google Scholar]
- Russello SV, Shore SK. SRC in human carcinogenesis. Front Biosci. 2004;9:139–144. doi: 10.2741/1138. [DOI] [PubMed] [Google Scholar]
- Sastry KS, Smith AJ, Karpova Y, Datta SR, Kulik G. Diverse antiapoptotic signaling pathways activated by vasoactive intestinal polypeptide, epidermal growth factor, and phosphatidylinositol 3-kinase in prostate cancer cells converge on BAD. J Biol Chem. 2006;281:20891–20901. doi: 10.1074/jbc.M602928200. [DOI] [PubMed] [Google Scholar]
- Scholz G, Martinerie C, Perbal B, Hanafusa H. Transcriptional down regulation of the nov proto-oncogene in fibroblasts transformed by p60v-src. Mol Cell Biol. 1996;16:481–486. doi: 10.1128/mcb.16.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Jia Z, Nagele RG, Ichikawa H, Goldberg GS. SRC uses Cas to suppress Fhl1 in order to promote nonanchored growth and migration of tumor cells. Cancer Res. 2006;66:1543–1552. doi: 10.1158/0008-5472.CAN-05-3152. [DOI] [PubMed] [Google Scholar]
- Shtivelman E, Cohen FE, Bishop JM. A human gene (AHNAK) encoding an unusually large protein with a 1.2-microns polyionic rod structure. Proc Natl Acad Sci U S A. 1992;89:5472–5476. doi: 10.1073/pnas.89.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stofega MR, Yu CL, Wu J, Jove R. Activation of extracellular signal-regulated kinase (ERK) by mitogenic stimuli is repressed in v-Src-transformed cells. Cell Growth Differ. 1997;8:113–119. [PubMed] [Google Scholar]
- Stove C, Vanrobaeys F, Devreese B, Van Beeumen J, Mareel M, Bracke M. Melanoma cells secrete follistatin, an antagonist of activin-mediated growth inhibition. Oncogene. 2004;23:5330–5339. doi: 10.1038/sj.onc.1207699. [DOI] [PubMed] [Google Scholar]
- Sudol M. Hormonal regulation of protein synthesis in cultured kidney cells. Mol Cell Endocrinol. 1985;40:245–255. doi: 10.1016/0303-7207(85)90180-7. [DOI] [PubMed] [Google Scholar]
- Sudol M, Greulich H, Newman L, Sarkar A, Sukegawa J, Yamamoto T. A novel Yes-related kinase, Yrk, is expressed at elevated levels in neural and hematopoietic tissues. Oncogene. 1993;8:823–831. [PubMed] [Google Scholar]
- Sugano S, Stoeckle MY, Hanafusa H. Transformation by Rous sarcoma virus induces a novel gene with homology to a mitogenic platelet protein. Cell. 1987;49:321–328. doi: 10.1016/0092-8674(87)90284-4. [DOI] [PubMed] [Google Scholar]
- Tian Q, Stepaniants SB, Mao M, Weng L, Feetham MC, Doyle MJ, Yi EC, Dai H, Thorsson V, Eng J, et al. Integrated genomic and proteomic analyses of gene expression in Mammalian cells. Mol Cell Proteomics. 2004;3:960–969. doi: 10.1074/mcp.M400055-MCP200. [DOI] [PubMed] [Google Scholar]
- Unkeless JC, Tobia A, Ossowski L, Quigley JP, Rifkin DB, Reich E. An enzymatic function associated with transformation of fibroblasts by oncogenic viruses. I Chick embryo fibroblast cultures transformed by avian RNA tumor viruses. J Exp Med. 1973;137:85–111. doi: 10.1084/jem.137.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyke T, Jacks T. Cancer modeling in the modern era: progress and challenges. Cell. 2002;108:135–144. doi: 10.1016/s0092-8674(02)00621-9. [DOI] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Vogt PK. the oncoprotein. Oncogene. 2001 Jun;20:2365–2377. doi: 10.1038/sj.onc.1204443. [DOI] [PubMed] [Google Scholar]
- Wallis JW, Aerts J, Groenen MA, Crooijmans RP, Layman D, Graves TA, Scheer DE, Kremitzki C, Fedele MJ, Mudd NK, et al. A physical map of the chicken genome. Nature. 2004;432:761–764. doi: 10.1038/nature03030. [DOI] [PubMed] [Google Scholar]
- Wang LH, Beckson M, Anderson SM, Hanafusa H. Identification of the viral sequence required for the generation of recovered avian sarcoma viruses and characterization of a series of replication-defective recovered avian sarcoma viruses. J Virol. 1984;49:881–891. doi: 10.1128/jvi.49.3.881-891.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MK, Rall TB, Weber MJ. Differential regulation of glucose transporter isoforms by the src oncogene in chicken embryo fibroblasts. Mol Cell Biol. 1991;11:4448–4454. doi: 10.1128/mcb.11.9.4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
To identify sets of genes on the basis of similarities in the pattern with which their expression changed in RSV transformed cells, we applied two-dimensional hierarchical clustering. We considered all the genes with p-value of less than 0.05 and the linear fold change more that 2.5. The Spotfire Software was then used to cluster the experiments (columns) and gene probes (rows) into a heat map using the un-weighted average and Euclidean distance methods.
List of genes and open reading frames that are induced (I) or repressed (D, after decrease) for equal or more than 2.5 fold (linear, in either direction) and with the p-value less than 0.05 in RSV-SR transformed CEFs compared to RSV-SR td106 mutant infected CEFs.
Summary of gene ontology terms associated with transcripts significantly modulated by v-src.


