Figure 2.
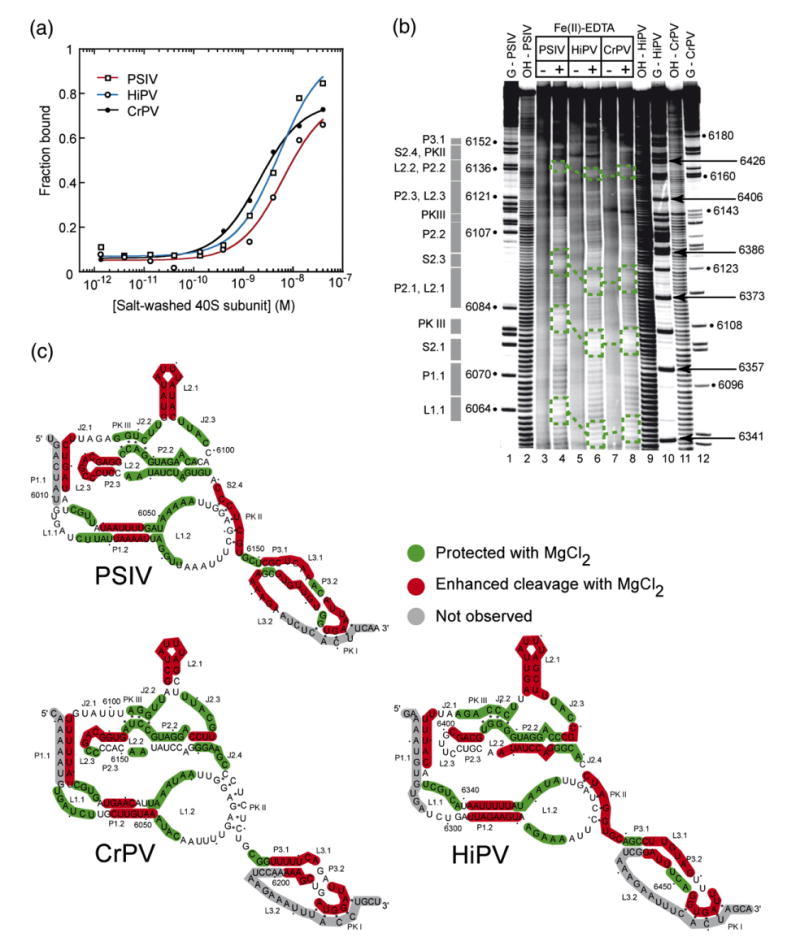
Small ribosomal subunit binding affinity comparison and structural comparison of class 1 IGR IRESes. (a) Binding isotherms of purified 40 S subunit to PSIV, CrPV, and HiPV IRES RNAs (a single experiment is shown) obtained from filter binding in a buffer containing 2.5 mM Mg2+ and 300 mM K+. These conditions minimize non-specific binding which occurs between purified ribosomes and RNA, and were chosen to match previous quantitative binding assays.27 (b) Example of a hydroxyl radical probing experiment of the PSIV (lanes 3 and 4), CrPV (lanes 7 and 8), and HiPV (lanes 5 and 6) IRES RNAs. A few examples of regions of the backbone protected in all three IRES RNAs are indicated with boxes. Repeating this experiment with different gel run times and quantitative analysis of the resulting gels led to a mapping of most of the RNA backbone. (c) Hydroxyl radical cleavage pattern for all three IRESes mapped onto their secondary structures.
